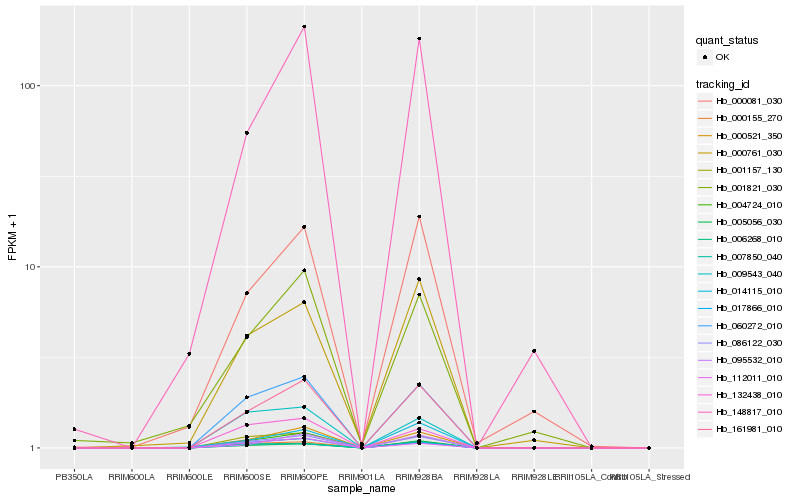
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_161981_010 |
0.0 |
- |
- |
brassinosteroid insensitive 1 -like protein [Gossypium arboreum] |
| 2 |
Hb_086122_030 |
0.0235295206 |
- |
- |
D-glycerate 3-kinase, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_060272_010 |
0.0677938345 |
- |
- |
PREDICTED: gibberellin 20 oxidase 2 [Populus euphratica] |
| 4 |
Hb_000521_350 |
0.0736216641 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_000155_270 |
0.0852671956 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF098 [Jatropha curcas] |
| 6 |
Hb_006268_010 |
0.1046077572 |
- |
- |
hypothetical protein EUGRSUZ_B00966 [Eucalyptus grandis] |
| 7 |
Hb_148817_010 |
0.1058607784 |
- |
- |
PREDICTED: CASP-like protein 1D1 [Jatropha curcas] |
| 8 |
Hb_001157_130 |
0.1071756101 |
- |
- |
PREDICTED: transmembrane protein 45B-like [Jatropha curcas] |
| 9 |
Hb_095532_010 |
0.1092085372 |
- |
- |
BnaC05g27570D [Brassica napus] |
| 10 |
Hb_005056_030 |
0.1221274199 |
- |
- |
DUF26 domain-containing protein 2 precursor, putative [Ricinus communis] |
| 11 |
Hb_017866_010 |
0.1234197567 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 12 |
Hb_000761_030 |
0.1242203955 |
transcription factor |
TF Family: ERF |
Transcriptional factor TINY, putative [Ricinus communis] |
| 13 |
Hb_000081_030 |
0.1276184214 |
- |
- |
BnaC09g05810D [Brassica napus] |
| 14 |
Hb_007850_040 |
0.1352996148 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_014115_010 |
0.1388201382 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_112011_010 |
0.1414252299 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_132438_010 |
0.1486358061 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 18 |
Hb_004724_010 |
0.1504505498 |
- |
- |
- |
| 19 |
Hb_001821_030 |
0.1514412316 |
- |
- |
hypothetical protein JCGZ_13357 [Jatropha curcas] |
| 20 |
Hb_009543_040 |
0.1545891782 |
- |
- |
- |