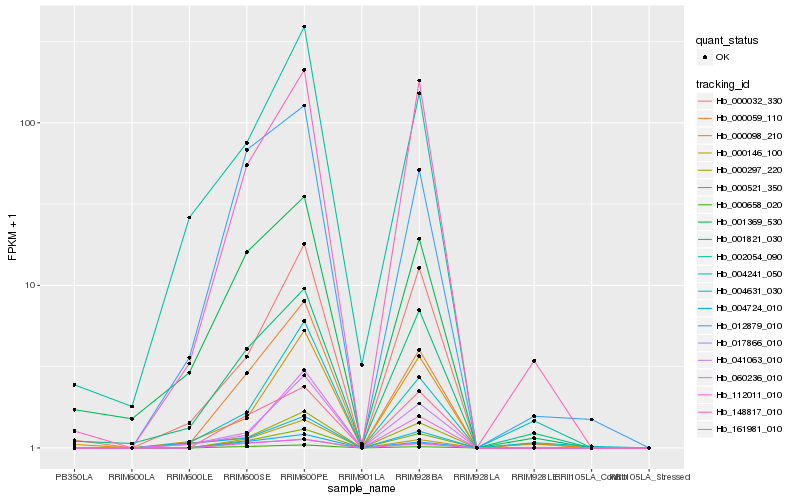
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000059_110 |
0.0 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus tremula x Populus tremuloides] |
| 2 |
Hb_000521_350 |
0.1122587677 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_004724_010 |
0.1122614848 |
- |
- |
- |
| 4 |
Hb_004241_050 |
0.1124452477 |
- |
- |
Indole-3-acetic acid-amido synthetase GH3.6, putative [Ricinus communis] |
| 5 |
Hb_000146_100 |
0.1132947037 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000658_020 |
0.1201774958 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 1 [Musa acuminata subsp. malaccensis] |
| 7 |
Hb_041063_010 |
0.124065106 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 8 |
Hb_112011_010 |
0.1272094178 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_012879_010 |
0.1296031792 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001821_030 |
0.1345335124 |
- |
- |
hypothetical protein JCGZ_13357 [Jatropha curcas] |
| 11 |
Hb_002054_090 |
0.1345931169 |
- |
- |
PREDICTED: 36.4 kDa proline-rich protein-like [Gossypium raimondii] |
| 12 |
Hb_148817_010 |
0.1392383024 |
- |
- |
PREDICTED: CASP-like protein 1D1 [Jatropha curcas] |
| 13 |
Hb_017866_010 |
0.1414413109 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 14 |
Hb_001369_530 |
0.1483801273 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 15 |
Hb_000032_330 |
0.1517654994 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000098_210 |
0.1526989447 |
- |
- |
PREDICTED: uncharacterized protein LOC105633345 [Jatropha curcas] |
| 17 |
Hb_004631_030 |
0.1546371404 |
- |
- |
PREDICTED: putative clathrin assembly protein At4g40080 [Jatropha curcas] |
| 18 |
Hb_161981_010 |
0.1549905792 |
- |
- |
brassinosteroid insensitive 1 -like protein [Gossypium arboreum] |
| 19 |
Hb_060236_010 |
0.1557015822 |
- |
- |
hypothetical protein JCGZ_26099 [Jatropha curcas] |
| 20 |
Hb_000297_220 |
0.1617851907 |
- |
- |
SAUR family protein [Theobroma cacao] |