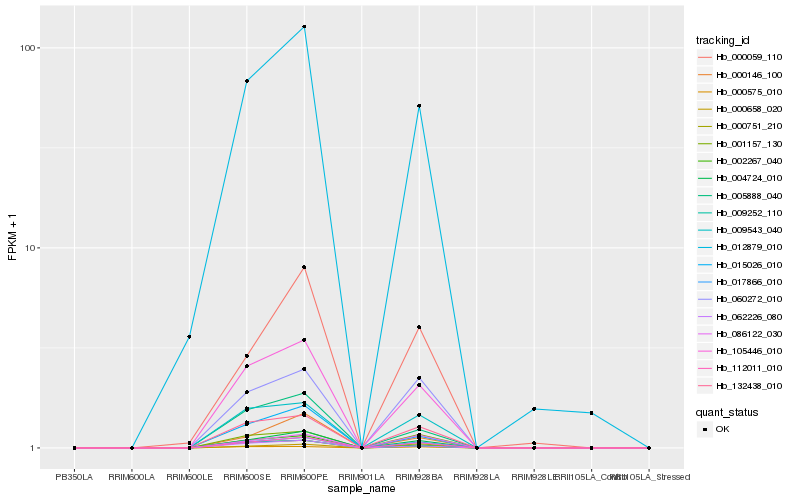
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_112011_010 |
0.0 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000658_020 |
0.0216648837 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 1 [Musa acuminata subsp. malaccensis] |
| 3 |
Hb_004724_010 |
0.0260194782 |
- |
- |
- |
| 4 |
Hb_017866_010 |
0.0312893818 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 5 |
Hb_105446_010 |
0.0466274076 |
- |
- |
- |
| 6 |
Hb_000751_210 |
0.0642187863 |
- |
- |
PREDICTED: petal death protein isoform X2 [Jatropha curcas] |
| 7 |
Hb_132438_010 |
0.0703611771 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 8 |
Hb_012879_010 |
0.0909847758 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_009543_040 |
0.0954357568 |
- |
- |
- |
| 10 |
Hb_001157_130 |
0.0972384085 |
- |
- |
PREDICTED: transmembrane protein 45B-like [Jatropha curcas] |
| 11 |
Hb_062226_080 |
0.1061420995 |
- |
- |
- |
| 12 |
Hb_060272_010 |
0.1063899718 |
- |
- |
PREDICTED: gibberellin 20 oxidase 2 [Populus euphratica] |
| 13 |
Hb_005888_040 |
0.1079917705 |
- |
- |
PREDICTED: gibberellin 20 oxidase 2 [Jatropha curcas] |
| 14 |
Hb_015026_010 |
0.1122516786 |
- |
- |
- |
| 15 |
Hb_000146_100 |
0.1260871828 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000059_110 |
0.1272094178 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus tremula x Populus tremuloides] |
| 17 |
Hb_002267_040 |
0.1299881926 |
- |
- |
- |
| 18 |
Hb_009252_110 |
0.1302367634 |
- |
- |
hypothetical protein JCGZ_11492 [Jatropha curcas] |
| 19 |
Hb_086122_030 |
0.1325888191 |
- |
- |
D-glycerate 3-kinase, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_000575_010 |
0.1347640891 |
- |
- |
PREDICTED: uncharacterized protein LOC103454030 [Malus domestica] |