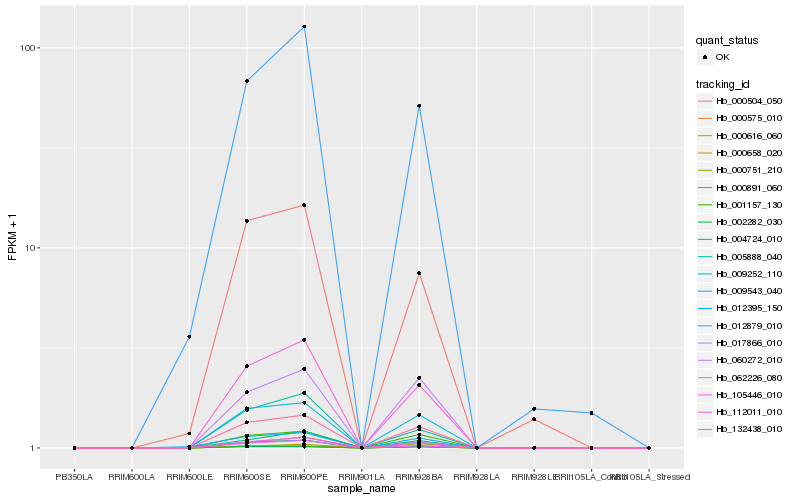
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000751_210 |
0.0 |
- |
- |
PREDICTED: petal death protein isoform X2 [Jatropha curcas] |
| 2 |
Hb_132438_010 |
0.0185000482 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 3 |
Hb_009543_040 |
0.0394745012 |
- |
- |
- |
| 4 |
Hb_105446_010 |
0.039774779 |
- |
- |
- |
| 5 |
Hb_017866_010 |
0.0489641039 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 6 |
Hb_112011_010 |
0.0642187863 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_001157_130 |
0.0718085931 |
- |
- |
PREDICTED: transmembrane protein 45B-like [Jatropha curcas] |
| 8 |
Hb_000658_020 |
0.081061742 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 1 [Musa acuminata subsp. malaccensis] |
| 9 |
Hb_000575_010 |
0.0825114997 |
- |
- |
PREDICTED: uncharacterized protein LOC103454030 [Malus domestica] |
| 10 |
Hb_000616_060 |
0.0858817854 |
- |
- |
- |
| 11 |
Hb_004724_010 |
0.0887388944 |
- |
- |
- |
| 12 |
Hb_000891_060 |
0.090460709 |
- |
- |
hypothetical protein JCGZ_11234 [Jatropha curcas] |
| 13 |
Hb_002282_030 |
0.0980704113 |
- |
- |
- |
| 14 |
Hb_060272_010 |
0.1032035503 |
- |
- |
PREDICTED: gibberellin 20 oxidase 2 [Populus euphratica] |
| 15 |
Hb_000504_050 |
0.1067409995 |
transcription factor |
TF Family: LOB |
LOB domain protein 25 [Populus trichocarpa] |
| 16 |
Hb_012879_010 |
0.1085866623 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005888_040 |
0.1091926131 |
- |
- |
PREDICTED: gibberellin 20 oxidase 2 [Jatropha curcas] |
| 18 |
Hb_062226_080 |
0.1122012765 |
- |
- |
- |
| 19 |
Hb_009252_110 |
0.1143013941 |
- |
- |
hypothetical protein JCGZ_11492 [Jatropha curcas] |
| 20 |
Hb_012395_150 |
0.1218484236 |
transcription factor |
TF Family: Orphans |
PREDICTED: two-component response regulator-like APRR7 [Jatropha curcas] |