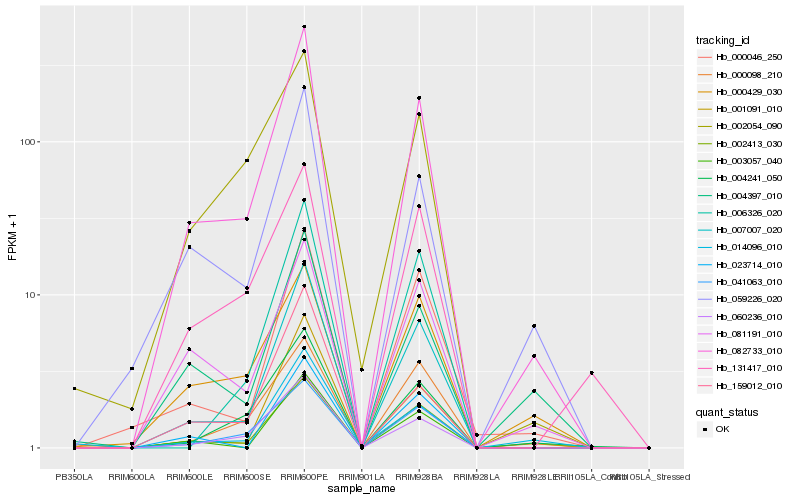
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_082733_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_23600 [Jatropha curcas] |
| 2 |
Hb_007007_020 |
0.0702456979 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002413_030 |
0.0835822647 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 4 |
Hb_060236_010 |
0.0918322606 |
- |
- |
hypothetical protein JCGZ_26099 [Jatropha curcas] |
| 5 |
Hb_059226_020 |
0.1091478281 |
- |
- |
hypothetical protein B456_008G287700 [Gossypium raimondii] |
| 6 |
Hb_041063_010 |
0.1200576223 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 7 |
Hb_001091_010 |
0.1234813667 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 isoform X2 [Malus domestica] |
| 8 |
Hb_004241_050 |
0.1238105808 |
- |
- |
Indole-3-acetic acid-amido synthetase GH3.6, putative [Ricinus communis] |
| 9 |
Hb_081191_010 |
0.1291435121 |
- |
- |
PREDICTED: pathogenesis-related protein PR-1-like [Jatropha curcas] |
| 10 |
Hb_004397_010 |
0.1354828873 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 11 |
Hb_159012_010 |
0.1363333369 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g06940 [Jatropha curcas] |
| 12 |
Hb_000046_250 |
0.1388178624 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003057_040 |
0.1440239861 |
- |
- |
PREDICTED: salt stress-induced hydrophobic peptide ESI3 [Vitis vinifera] |
| 14 |
Hb_002054_090 |
0.1468194445 |
- |
- |
PREDICTED: 36.4 kDa proline-rich protein-like [Gossypium raimondii] |
| 15 |
Hb_014096_010 |
0.1484372194 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 16 |
Hb_131417_010 |
0.1488641071 |
- |
- |
hypothetical protein POPTR_0011s066602g, partial [Populus trichocarpa] |
| 17 |
Hb_006326_020 |
0.1510456087 |
- |
- |
- |
| 18 |
Hb_023714_010 |
0.1519177633 |
- |
- |
hypothetical protein B456_007G377100 [Gossypium raimondii] |
| 19 |
Hb_000098_210 |
0.1559329757 |
- |
- |
PREDICTED: uncharacterized protein LOC105633345 [Jatropha curcas] |
| 20 |
Hb_000429_030 |
0.1569475009 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |