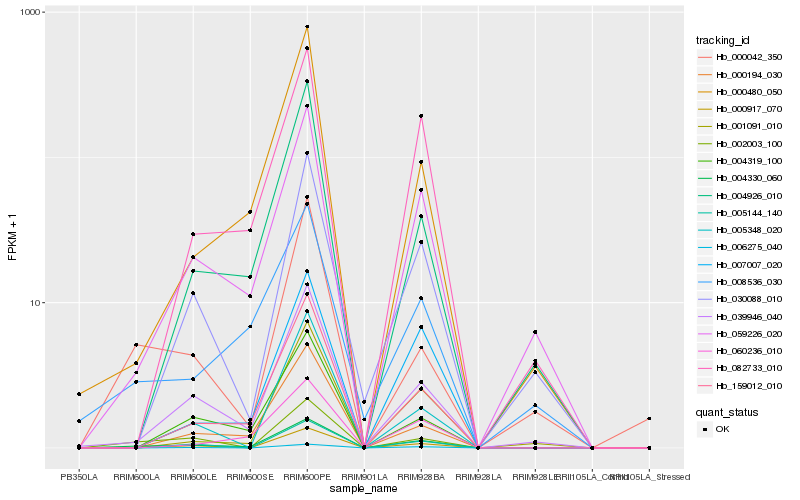
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004330_060 |
0.0 |
- |
- |
hypothetical protein POPTR_0011s01530g [Populus trichocarpa] |
| 2 |
Hb_059226_020 |
0.1315908799 |
- |
- |
hypothetical protein B456_008G287700 [Gossypium raimondii] |
| 3 |
Hb_039946_040 |
0.1336740312 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 4 |
Hb_159012_010 |
0.1508190669 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g06940 [Jatropha curcas] |
| 5 |
Hb_002003_100 |
0.1575388042 |
- |
- |
- |
| 6 |
Hb_000480_050 |
0.1693893497 |
- |
- |
hypothetical protein PHAVU_004G154200g [Phaseolus vulgaris] |
| 7 |
Hb_082733_010 |
0.1695234532 |
- |
- |
hypothetical protein JCGZ_23600 [Jatropha curcas] |
| 8 |
Hb_000194_030 |
0.1697294695 |
transcription factor |
TF Family: NAC |
NAC transcription factor 059 [Jatropha curcas] |
| 9 |
Hb_004926_010 |
0.1737157879 |
- |
- |
hypothetical protein JCGZ_27059 [Jatropha curcas] |
| 10 |
Hb_004319_100 |
0.175876869 |
- |
- |
PREDICTED: gibberellin-regulated protein 14 isoform X3 [Jatropha curcas] |
| 11 |
Hb_000917_070 |
0.1792885421 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_008536_030 |
0.1796304294 |
- |
- |
hypothetical protein POPTR_0005s08290g [Populus trichocarpa] |
| 13 |
Hb_005144_140 |
0.1807405942 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_007007_020 |
0.1822461817 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000042_350 |
0.1849311813 |
- |
- |
hypothetical protein JCGZ_00710 [Jatropha curcas] |
| 16 |
Hb_060236_010 |
0.1877144286 |
- |
- |
hypothetical protein JCGZ_26099 [Jatropha curcas] |
| 17 |
Hb_001091_010 |
0.1906418845 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 isoform X2 [Malus domestica] |
| 18 |
Hb_006275_040 |
0.191127499 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 19 |
Hb_030088_010 |
0.1918064517 |
- |
- |
basic blue copper family protein [Populus trichocarpa] |
| 20 |
Hb_005348_020 |
0.1940901859 |
- |
- |
conserved hypothetical protein [Ricinus communis] |