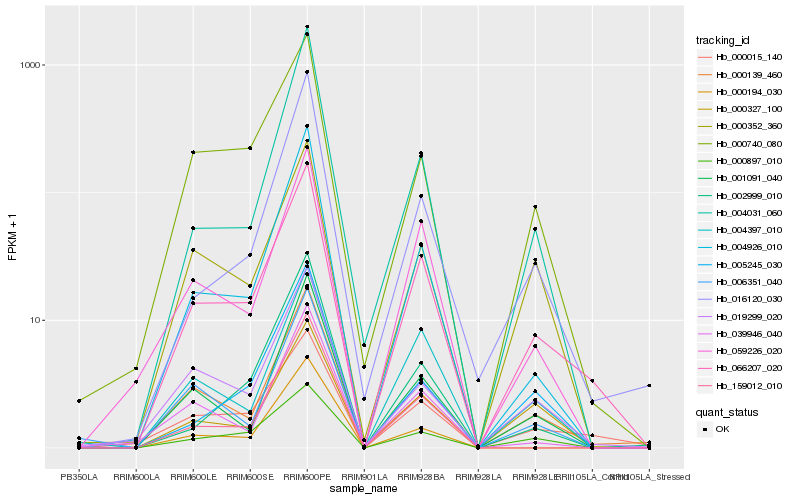
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_066207_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_22048 [Jatropha curcas] |
| 2 |
Hb_000015_140 |
0.1070143792 |
- |
- |
PREDICTED: CASP-like protein 4C1 [Vitis vinifera] |
| 3 |
Hb_059226_020 |
0.1229403529 |
- |
- |
hypothetical protein B456_008G287700 [Gossypium raimondii] |
| 4 |
Hb_000740_080 |
0.1272380714 |
- |
- |
- |
| 5 |
Hb_000897_010 |
0.1274777306 |
- |
- |
hypothetical protein JCGZ_13196 [Jatropha curcas] |
| 6 |
Hb_004397_010 |
0.1298767621 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 7 |
Hb_004926_010 |
0.1316774933 |
- |
- |
hypothetical protein JCGZ_27059 [Jatropha curcas] |
| 8 |
Hb_000139_460 |
0.1325788988 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 9 |
Hb_001091_040 |
0.1379242573 |
- |
- |
hypothetical protein JCGZ_01204 [Jatropha curcas] |
| 10 |
Hb_019299_020 |
0.1396407396 |
- |
- |
PREDICTED: putative kinase-like protein TMKL1 [Jatropha curcas] |
| 11 |
Hb_000352_360 |
0.1397918943 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 1-like [Jatropha curcas] |
| 12 |
Hb_006351_040 |
0.1427830697 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000327_100 |
0.1431852982 |
- |
- |
hypothetical protein JCGZ_06658 [Jatropha curcas] |
| 14 |
Hb_159012_010 |
0.1541203084 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g06940 [Jatropha curcas] |
| 15 |
Hb_039946_040 |
0.1541607682 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 16 |
Hb_004031_060 |
0.1547670646 |
- |
- |
hypothetical protein B456_007G3219002, partial [Gossypium raimondii] |
| 17 |
Hb_016120_030 |
0.1556574631 |
- |
- |
hypothetical protein EUGRSUZ_D00080 [Eucalyptus grandis] |
| 18 |
Hb_005245_030 |
0.156859115 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At3g07570 [Jatropha curcas] |
| 19 |
Hb_002999_010 |
0.1598052774 |
- |
- |
PREDICTED: extracellular ribonuclease LE-like [Vitis vinifera] |
| 20 |
Hb_000194_030 |
0.1614835738 |
transcription factor |
TF Family: NAC |
NAC transcription factor 059 [Jatropha curcas] |