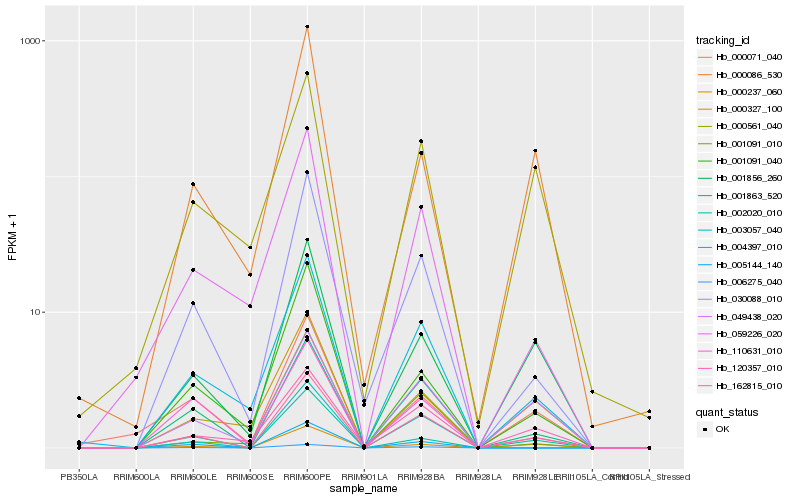
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_162815_010 |
0.0 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 2 |
Hb_001863_520 |
0.0949240774 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_030088_010 |
0.1213631397 |
- |
- |
basic blue copper family protein [Populus trichocarpa] |
| 4 |
Hb_049438_020 |
0.1219732062 |
- |
- |
PREDICTED: peroxidase 20 [Jatropha curcas] |
| 5 |
Hb_004397_010 |
0.1219903985 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 6 |
Hb_001856_260 |
0.1299372307 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 7 |
Hb_120357_010 |
0.1446239623 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 8 |
Hb_000237_060 |
0.1467776733 |
- |
- |
hypothetical protein PRUPE_ppa022052mg, partial [Prunus persica] |
| 9 |
Hb_002020_010 |
0.1624290664 |
- |
- |
kinase-like protein [Corylus avellana] |
| 10 |
Hb_001091_040 |
0.1627785949 |
- |
- |
hypothetical protein JCGZ_01204 [Jatropha curcas] |
| 11 |
Hb_059226_020 |
0.1632661755 |
- |
- |
hypothetical protein B456_008G287700 [Gossypium raimondii] |
| 12 |
Hb_110631_010 |
0.1642320578 |
- |
- |
hypothetical protein JCGZ_04359 [Jatropha curcas] |
| 13 |
Hb_006275_040 |
0.1648150465 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 14 |
Hb_000327_100 |
0.1703239007 |
- |
- |
hypothetical protein JCGZ_06658 [Jatropha curcas] |
| 15 |
Hb_003057_040 |
0.1715412471 |
- |
- |
PREDICTED: salt stress-induced hydrophobic peptide ESI3 [Vitis vinifera] |
| 16 |
Hb_000086_530 |
0.1732227765 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001091_010 |
0.175174284 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 isoform X2 [Malus domestica] |
| 18 |
Hb_005144_140 |
0.1764197946 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000561_040 |
0.1774407481 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |
| 20 |
Hb_000071_040 |
0.1783268839 |
- |
- |
hypothetical protein POPTR_0001s24580g [Populus trichocarpa] |