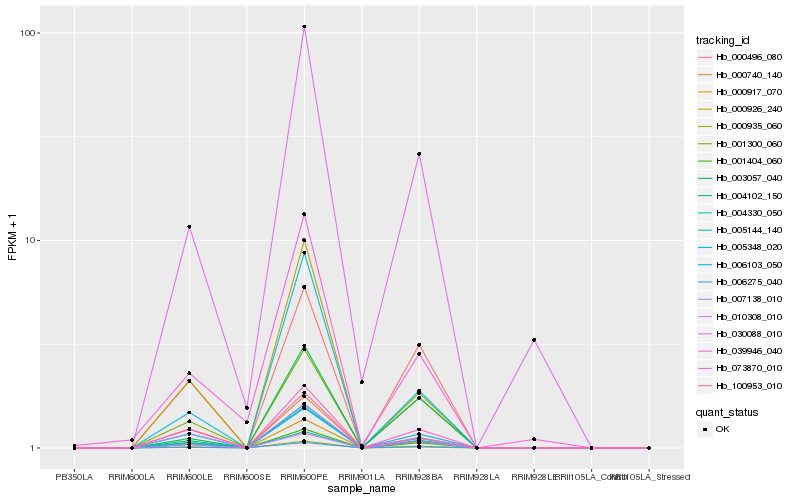
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005144_140 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000917_070 |
0.0170879385 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004102_150 |
0.0627651636 |
- |
- |
Late embryogenesis abundant protein D-7, putative [Ricinus communis] |
| 4 |
Hb_006275_040 |
0.0708703284 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 5 |
Hb_000935_060 |
0.0733289169 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 8-like [Jatropha curcas] |
| 6 |
Hb_073870_010 |
0.094786445 |
- |
- |
hypothetical protein RCOM_0725510 [Ricinus communis] |
| 7 |
Hb_005348_020 |
0.097553813 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001404_060 |
0.1113758032 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_000926_240 |
0.119434191 |
- |
- |
hypothetical protein PRUPE_ppb018041mg [Prunus persica] |
| 10 |
Hb_030088_010 |
0.1199700859 |
- |
- |
basic blue copper family protein [Populus trichocarpa] |
| 11 |
Hb_000740_140 |
0.1210168228 |
transcription factor |
TF Family: OFP |
PREDICTED: probable transcription repressor OFP9 [Jatropha curcas] |
| 12 |
Hb_003057_040 |
0.1239632444 |
- |
- |
PREDICTED: salt stress-induced hydrophobic peptide ESI3 [Vitis vinifera] |
| 13 |
Hb_001300_060 |
0.1252954129 |
- |
- |
PREDICTED: monothiol glutaredoxin-S6-like [Jatropha curcas] |
| 14 |
Hb_007138_010 |
0.13094266 |
- |
- |
PREDICTED: putative laccase-9 [Jatropha curcas] |
| 15 |
Hb_006103_050 |
0.1357226911 |
- |
- |
- |
| 16 |
Hb_004330_050 |
0.1378827291 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_010308_010 |
0.1397358626 |
- |
- |
hypothetical protein JCGZ_22818 [Jatropha curcas] |
| 18 |
Hb_000496_080 |
0.1408951326 |
- |
- |
Proteinase inhibitor, putative [Ricinus communis] |
| 19 |
Hb_039946_040 |
0.1420625999 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 20 |
Hb_100953_010 |
0.1438339523 |
- |
- |
hypothetical protein JCGZ_10310 [Jatropha curcas] |