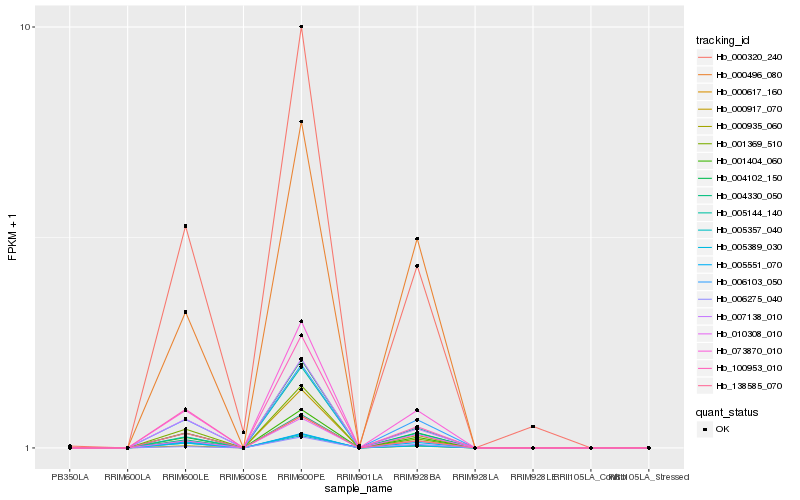
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_073870_010 |
0.0 |
- |
- |
hypothetical protein RCOM_0725510 [Ricinus communis] |
| 2 |
Hb_001404_060 |
0.0215353996 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_006103_050 |
0.0485791145 |
- |
- |
- |
| 4 |
Hb_007138_010 |
0.0528080339 |
- |
- |
PREDICTED: putative laccase-9 [Jatropha curcas] |
| 5 |
Hb_100953_010 |
0.0761291998 |
- |
- |
hypothetical protein JCGZ_10310 [Jatropha curcas] |
| 6 |
Hb_000935_060 |
0.0901410492 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 8-like [Jatropha curcas] |
| 7 |
Hb_001369_510 |
0.0910880108 |
- |
- |
PREDICTED: uncharacterized protein LOC105766768 [Gossypium raimondii] |
| 8 |
Hb_004102_150 |
0.0938068115 |
- |
- |
Late embryogenesis abundant protein D-7, putative [Ricinus communis] |
| 9 |
Hb_005389_030 |
0.0945941644 |
- |
- |
hypothetical protein VITISV_023348 [Vitis vinifera] |
| 10 |
Hb_005144_140 |
0.094786445 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000320_240 |
0.1018423857 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 12 |
Hb_005357_040 |
0.102027945 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 13 |
Hb_004330_050 |
0.1034551743 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000917_070 |
0.1074546176 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005551_070 |
0.1082244081 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 16 |
Hb_010308_010 |
0.1084835187 |
- |
- |
hypothetical protein JCGZ_22818 [Jatropha curcas] |
| 17 |
Hb_000496_080 |
0.109431014 |
- |
- |
Proteinase inhibitor, putative [Ricinus communis] |
| 18 |
Hb_138585_070 |
0.1127768666 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_006275_040 |
0.1131904839 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 20 |
Hb_000617_160 |
0.1193799692 |
- |
- |
- |