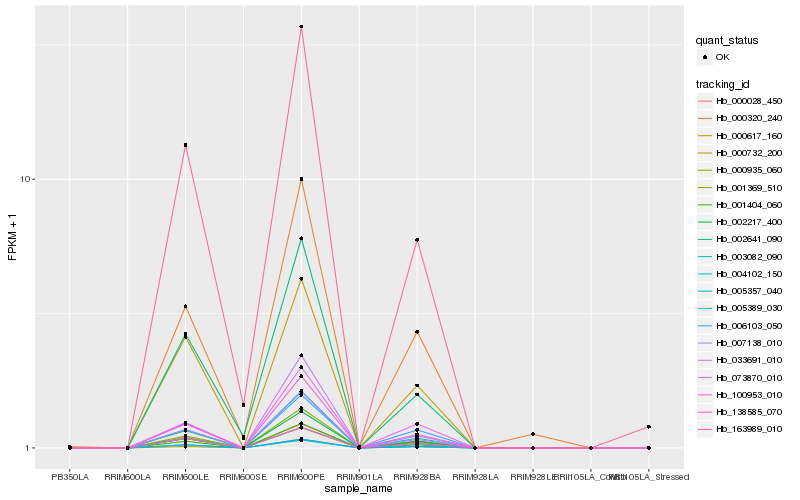
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007138_010 |
0.0 |
- |
- |
PREDICTED: putative laccase-9 [Jatropha curcas] |
| 2 |
Hb_100953_010 |
0.0251936822 |
- |
- |
hypothetical protein JCGZ_10310 [Jatropha curcas] |
| 3 |
Hb_001369_510 |
0.0434108852 |
- |
- |
PREDICTED: uncharacterized protein LOC105766768 [Gossypium raimondii] |
| 4 |
Hb_005389_030 |
0.045817252 |
- |
- |
hypothetical protein VITISV_023348 [Vitis vinifera] |
| 5 |
Hb_073870_010 |
0.0528080339 |
- |
- |
hypothetical protein RCOM_0725510 [Ricinus communis] |
| 6 |
Hb_001404_060 |
0.0602120437 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_005357_040 |
0.0619065673 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 8 |
Hb_006103_050 |
0.0748790038 |
- |
- |
- |
| 9 |
Hb_000028_450 |
0.076176872 |
- |
- |
PREDICTED: monoglyceride lipase-like [Jatropha curcas] |
| 10 |
Hb_138585_070 |
0.0799104997 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002217_400 |
0.0861479251 |
- |
- |
polynucleotidyl transferase, ribonuclease H-like protein [Arabidopsis thaliana] |
| 12 |
Hb_000617_160 |
0.0865972753 |
- |
- |
- |
| 13 |
Hb_000935_060 |
0.0932503611 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 8-like [Jatropha curcas] |
| 14 |
Hb_163989_010 |
0.0950380093 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003082_090 |
0.0952887833 |
- |
- |
hypothetical protein POPTR_0013s099451g, partial [Populus trichocarpa] |
| 16 |
Hb_000320_240 |
0.0954068687 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 17 |
Hb_000732_200 |
0.1020405489 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004102_150 |
0.102872295 |
- |
- |
Late embryogenesis abundant protein D-7, putative [Ricinus communis] |
| 19 |
Hb_002641_090 |
0.1056782958 |
- |
- |
PREDICTED: uncharacterized protein LOC105643559 [Jatropha curcas] |
| 20 |
Hb_033691_010 |
0.1198098964 |
- |
- |
Lignin-forming anionic peroxidase precursor, putative [Ricinus communis] |