| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
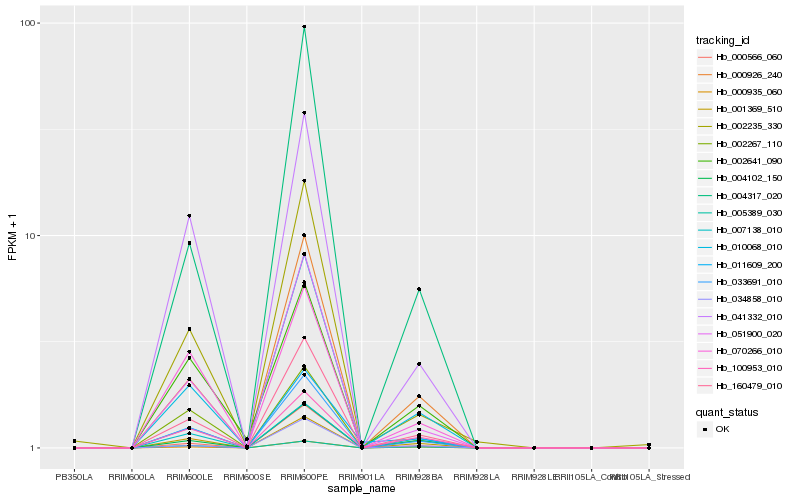
Hb_033691_010 |
0.0 |
- |
- |
Lignin-forming anionic peroxidase precursor, putative [Ricinus communis] |
| 2 |
Hb_160479_010 |
0.0350176496 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 3 |
Hb_010068_010 |
0.0565539838 |
- |
- |
PREDICTED: uncharacterized protein LOC105633273 [Jatropha curcas] |
| 4 |
Hb_051900_020 |
0.0668657101 |
- |
- |
JHL22C18.7 [Jatropha curcas] |
| 5 |
Hb_000926_240 |
0.0768325738 |
- |
- |
hypothetical protein PRUPE_ppb018041mg [Prunus persica] |
| 6 |
Hb_001369_510 |
0.0806177716 |
- |
- |
PREDICTED: uncharacterized protein LOC105766768 [Gossypium raimondii] |
| 7 |
Hb_041332_010 |
0.0836831807 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_100953_010 |
0.09915 |
- |
- |
hypothetical protein JCGZ_10310 [Jatropha curcas] |
| 9 |
Hb_000935_060 |
0.0993175405 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 8-like [Jatropha curcas] |
| 10 |
Hb_002267_110 |
0.1002777932 |
- |
- |
- |
| 11 |
Hb_002235_330 |
0.1056281251 |
- |
- |
PREDICTED: expansin-A1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004102_150 |
0.1102971033 |
- |
- |
Late embryogenesis abundant protein D-7, putative [Ricinus communis] |
| 13 |
Hb_070266_010 |
0.1114920656 |
- |
- |
LRR.XII-like protein, partial [Platanus x acerifolia] |
| 14 |
Hb_004317_020 |
0.112499743 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7-like [Malus domestica] |
| 15 |
Hb_000566_060 |
0.1162441128 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_007138_010 |
0.1198098964 |
- |
- |
PREDICTED: putative laccase-9 [Jatropha curcas] |
| 17 |
Hb_005389_030 |
0.1272910012 |
- |
- |
hypothetical protein VITISV_023348 [Vitis vinifera] |
| 18 |
Hb_002641_090 |
0.1278914625 |
- |
- |
PREDICTED: uncharacterized protein LOC105643559 [Jatropha curcas] |
| 19 |
Hb_034858_010 |
0.1364144358 |
- |
- |
- |
| 20 |
Hb_011609_200 |
0.1366093772 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8-like isoform X3 [Populus euphratica] |