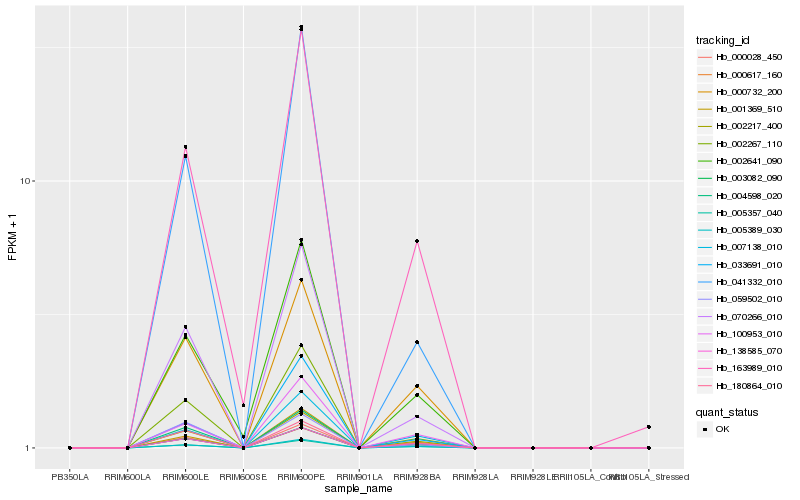
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_070266_010 |
0.0 |
- |
- |
LRR.XII-like protein, partial [Platanus x acerifolia] |
| 2 |
Hb_002267_110 |
0.0147508695 |
- |
- |
- |
| 3 |
Hb_041332_010 |
0.0521903213 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_004598_020 |
0.0581693055 |
transcription factor |
TF Family: ERF |
Transcriptional factor TINY, putative [Ricinus communis] |
| 5 |
Hb_003082_090 |
0.0763646548 |
- |
- |
hypothetical protein POPTR_0013s099451g, partial [Populus trichocarpa] |
| 6 |
Hb_002217_400 |
0.0968936908 |
- |
- |
polynucleotidyl transferase, ribonuclease H-like protein [Arabidopsis thaliana] |
| 7 |
Hb_000028_450 |
0.0970196448 |
- |
- |
PREDICTED: monoglyceride lipase-like [Jatropha curcas] |
| 8 |
Hb_001369_510 |
0.0998590845 |
- |
- |
PREDICTED: uncharacterized protein LOC105766768 [Gossypium raimondii] |
| 9 |
Hb_005389_030 |
0.1000668876 |
- |
- |
hypothetical protein VITISV_023348 [Vitis vinifera] |
| 10 |
Hb_002641_090 |
0.1073020581 |
- |
- |
PREDICTED: uncharacterized protein LOC105643559 [Jatropha curcas] |
| 11 |
Hb_100953_010 |
0.1103772647 |
- |
- |
hypothetical protein JCGZ_10310 [Jatropha curcas] |
| 12 |
Hb_033691_010 |
0.1114920656 |
- |
- |
Lignin-forming anionic peroxidase precursor, putative [Ricinus communis] |
| 13 |
Hb_005357_040 |
0.1131115853 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 14 |
Hb_163989_010 |
0.1137047346 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_180864_010 |
0.1217960544 |
- |
- |
hypothetical protein B456_010G097000 [Gossypium raimondii] |
| 16 |
Hb_059502_010 |
0.1256686776 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |
| 17 |
Hb_138585_070 |
0.12733923 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000617_160 |
0.1278439136 |
- |
- |
- |
| 19 |
Hb_007138_010 |
0.1328068445 |
- |
- |
PREDICTED: putative laccase-9 [Jatropha curcas] |
| 20 |
Hb_000732_200 |
0.1341778592 |
- |
- |
conserved hypothetical protein [Ricinus communis] |