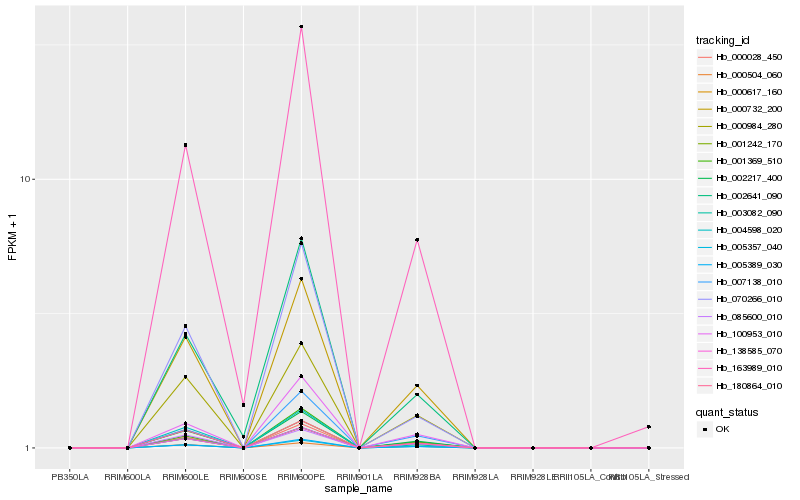
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000028_450 |
0.0 |
- |
- |
PREDICTED: monoglyceride lipase-like [Jatropha curcas] |
| 2 |
Hb_002217_400 |
0.0100614698 |
- |
- |
polynucleotidyl transferase, ribonuclease H-like protein [Arabidopsis thaliana] |
| 3 |
Hb_005357_040 |
0.0204898851 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 4 |
Hb_003082_090 |
0.0259929551 |
- |
- |
hypothetical protein POPTR_0013s099451g, partial [Populus trichocarpa] |
| 5 |
Hb_138585_070 |
0.0310737175 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_005389_030 |
0.0319840182 |
- |
- |
hypothetical protein VITISV_023348 [Vitis vinifera] |
| 7 |
Hb_000617_160 |
0.0325796293 |
- |
- |
- |
| 8 |
Hb_000732_200 |
0.0432868296 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000984_280 |
0.0676705774 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 2 [Jatropha curcas] |
| 10 |
Hb_100953_010 |
0.0707849854 |
- |
- |
hypothetical protein JCGZ_10310 [Jatropha curcas] |
| 11 |
Hb_004598_020 |
0.0761408415 |
transcription factor |
TF Family: ERF |
Transcriptional factor TINY, putative [Ricinus communis] |
| 12 |
Hb_007138_010 |
0.076176872 |
- |
- |
PREDICTED: putative laccase-9 [Jatropha curcas] |
| 13 |
Hb_180864_010 |
0.0772223595 |
- |
- |
hypothetical protein B456_010G097000 [Gossypium raimondii] |
| 14 |
Hb_001242_170 |
0.0795377424 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001369_510 |
0.0800532071 |
- |
- |
PREDICTED: uncharacterized protein LOC105766768 [Gossypium raimondii] |
| 16 |
Hb_163989_010 |
0.0855169909 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_085600_010 |
0.0955207805 |
- |
- |
PREDICTED: peroxidase 24-like [Jatropha curcas] |
| 18 |
Hb_070266_010 |
0.0970196448 |
- |
- |
LRR.XII-like protein, partial [Platanus x acerifolia] |
| 19 |
Hb_002641_090 |
0.0988968811 |
- |
- |
PREDICTED: uncharacterized protein LOC105643559 [Jatropha curcas] |
| 20 |
Hb_000504_060 |
0.106252441 |
- |
- |
Cell division cycle protein cdt2, putative [Ricinus communis] |