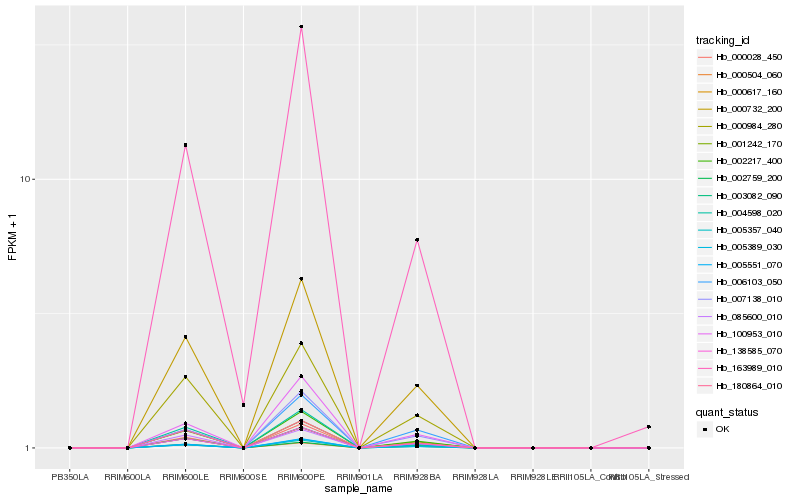
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000732_200 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000617_160 |
0.0155646784 |
- |
- |
- |
| 3 |
Hb_138585_070 |
0.0222285324 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000984_280 |
0.0329603314 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 2 [Jatropha curcas] |
| 5 |
Hb_002217_400 |
0.0390859208 |
- |
- |
polynucleotidyl transferase, ribonuclease H-like protein [Arabidopsis thaliana] |
| 6 |
Hb_005357_040 |
0.0427725841 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 7 |
Hb_000028_450 |
0.0432868296 |
- |
- |
PREDICTED: monoglyceride lipase-like [Jatropha curcas] |
| 8 |
Hb_001242_170 |
0.0541273349 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003082_090 |
0.0587609407 |
- |
- |
hypothetical protein POPTR_0013s099451g, partial [Populus trichocarpa] |
| 10 |
Hb_000504_060 |
0.0637156758 |
- |
- |
Cell division cycle protein cdt2, putative [Ricinus communis] |
| 11 |
Hb_180864_010 |
0.06498776 |
- |
- |
hypothetical protein B456_010G097000 [Gossypium raimondii] |
| 12 |
Hb_085600_010 |
0.0669109668 |
- |
- |
PREDICTED: peroxidase 24-like [Jatropha curcas] |
| 13 |
Hb_005389_030 |
0.067954208 |
- |
- |
hypothetical protein VITISV_023348 [Vitis vinifera] |
| 14 |
Hb_002759_200 |
0.0909187762 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 2 [Jatropha curcas] |
| 15 |
Hb_004598_020 |
0.0984712934 |
transcription factor |
TF Family: ERF |
Transcriptional factor TINY, putative [Ricinus communis] |
| 16 |
Hb_005551_070 |
0.0995914426 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 17 |
Hb_007138_010 |
0.1020405489 |
- |
- |
PREDICTED: putative laccase-9 [Jatropha curcas] |
| 18 |
Hb_100953_010 |
0.1054597267 |
- |
- |
hypothetical protein JCGZ_10310 [Jatropha curcas] |
| 19 |
Hb_163989_010 |
0.1097943088 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_006103_050 |
0.1098900767 |
- |
- |
- |