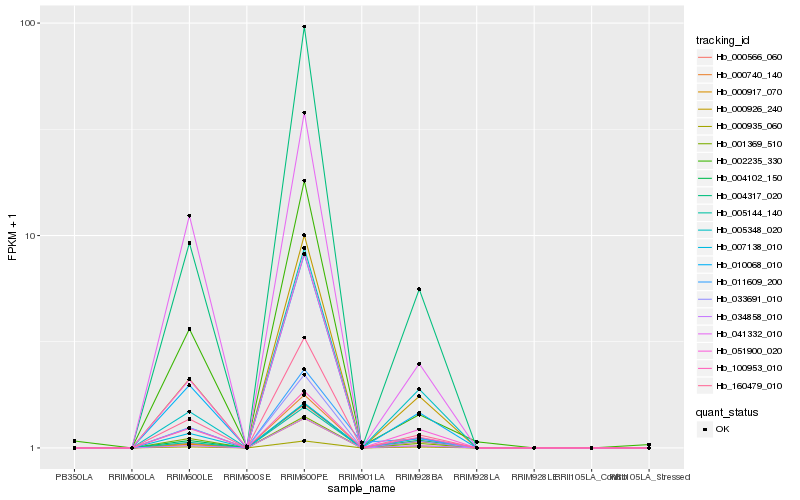
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010068_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633273 [Jatropha curcas] |
| 2 |
Hb_160479_010 |
0.0217440408 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 3 |
Hb_000926_240 |
0.0291161912 |
- |
- |
hypothetical protein PRUPE_ppb018041mg [Prunus persica] |
| 4 |
Hb_033691_010 |
0.0565539838 |
- |
- |
Lignin-forming anionic peroxidase precursor, putative [Ricinus communis] |
| 5 |
Hb_004317_020 |
0.0605292289 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7-like [Malus domestica] |
| 6 |
Hb_051900_020 |
0.0634544895 |
- |
- |
JHL22C18.7 [Jatropha curcas] |
| 7 |
Hb_000566_060 |
0.0686924718 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_034858_010 |
0.0808905826 |
- |
- |
- |
| 9 |
Hb_000935_060 |
0.0813939177 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 8-like [Jatropha curcas] |
| 10 |
Hb_004102_150 |
0.0870346845 |
- |
- |
Late embryogenesis abundant protein D-7, putative [Ricinus communis] |
| 11 |
Hb_002235_330 |
0.105293343 |
- |
- |
PREDICTED: expansin-A1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_005348_020 |
0.112546929 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000740_140 |
0.1155854391 |
transcription factor |
TF Family: OFP |
PREDICTED: probable transcription repressor OFP9 [Jatropha curcas] |
| 14 |
Hb_001369_510 |
0.1158827885 |
- |
- |
PREDICTED: uncharacterized protein LOC105766768 [Gossypium raimondii] |
| 15 |
Hb_100953_010 |
0.1307923887 |
- |
- |
hypothetical protein JCGZ_10310 [Jatropha curcas] |
| 16 |
Hb_000917_070 |
0.1368793218 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_041332_010 |
0.1374443955 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_011609_200 |
0.1411646707 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8-like isoform X3 [Populus euphratica] |
| 19 |
Hb_007138_010 |
0.1445182449 |
- |
- |
PREDICTED: putative laccase-9 [Jatropha curcas] |
| 20 |
Hb_005144_140 |
0.1461470401 |
- |
- |
conserved hypothetical protein [Ricinus communis] |