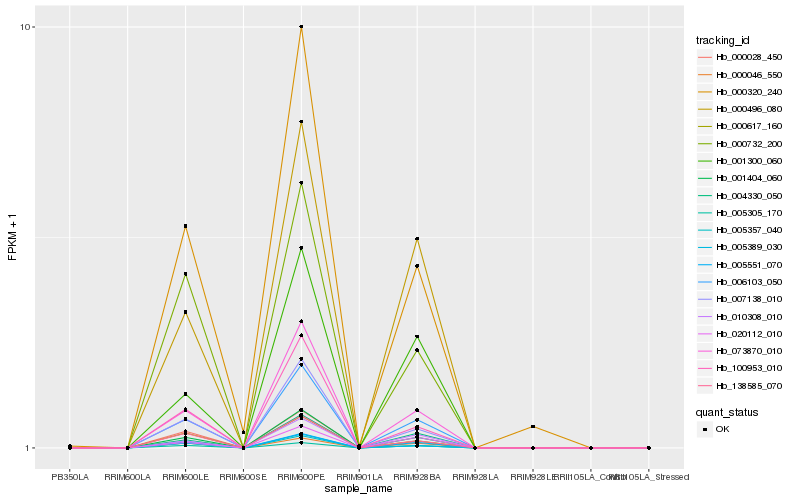
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006103_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_001404_060 |
0.0271616378 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_073870_010 |
0.0485791145 |
- |
- |
hypothetical protein RCOM_0725510 [Ricinus communis] |
| 4 |
Hb_005551_070 |
0.059993121 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 5 |
Hb_007138_010 |
0.0748790038 |
- |
- |
PREDICTED: putative laccase-9 [Jatropha curcas] |
| 6 |
Hb_004330_050 |
0.0839772249 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_010308_010 |
0.0896726932 |
- |
- |
hypothetical protein JCGZ_22818 [Jatropha curcas] |
| 8 |
Hb_000496_080 |
0.0902254579 |
- |
- |
Proteinase inhibitor, putative [Ricinus communis] |
| 9 |
Hb_005357_040 |
0.0933036397 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 10 |
Hb_138585_070 |
0.0942467319 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005305_170 |
0.0945453785 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 8 [Jatropha curcas] |
| 12 |
Hb_020112_010 |
0.0984879503 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 13 |
Hb_005389_030 |
0.0986043831 |
- |
- |
hypothetical protein VITISV_023348 [Vitis vinifera] |
| 14 |
Hb_100953_010 |
0.0993002406 |
- |
- |
hypothetical protein JCGZ_10310 [Jatropha curcas] |
| 15 |
Hb_000617_160 |
0.0996497174 |
- |
- |
- |
| 16 |
Hb_000046_550 |
0.1049765059 |
- |
- |
PREDICTED: protein ALWAYS EARLY 3 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000732_200 |
0.1098900767 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001300_060 |
0.1130751865 |
- |
- |
PREDICTED: monothiol glutaredoxin-S6-like [Jatropha curcas] |
| 19 |
Hb_000320_240 |
0.1135957482 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 20 |
Hb_000028_450 |
0.1136190126 |
- |
- |
PREDICTED: monoglyceride lipase-like [Jatropha curcas] |