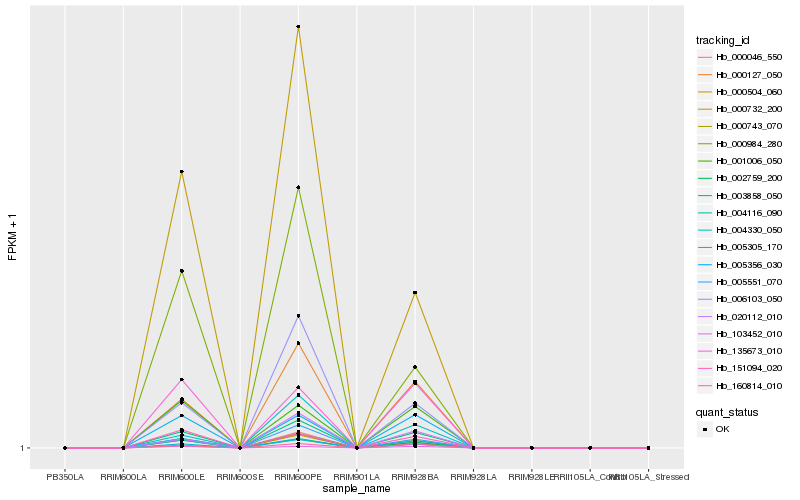
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003858_050 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa021882mg [Prunus persica] |
| 2 |
Hb_000046_550 |
0.031950251 |
- |
- |
PREDICTED: protein ALWAYS EARLY 3 isoform X2 [Jatropha curcas] |
| 3 |
Hb_005305_170 |
0.0432750861 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 8 [Jatropha curcas] |
| 4 |
Hb_000127_050 |
0.0630320909 |
- |
- |
- |
| 5 |
Hb_002759_200 |
0.0693012641 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 2 [Jatropha curcas] |
| 6 |
Hb_004116_090 |
0.072954827 |
- |
- |
- |
| 7 |
Hb_005551_070 |
0.0776570498 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000504_060 |
0.1088730298 |
- |
- |
Cell division cycle protein cdt2, putative [Ricinus communis] |
| 9 |
Hb_000743_070 |
0.1120827798 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21060 [Jatropha curcas] |
| 10 |
Hb_160814_010 |
0.1197235784 |
- |
- |
PREDICTED: probable sulfate transporter 3.5 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005356_030 |
0.122490041 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 12 |
Hb_103452_010 |
0.1276670503 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 13 |
Hb_001006_050 |
0.130470439 |
- |
- |
PREDICTED: uncharacterized protein LOC101777526 [Setaria italica] |
| 14 |
Hb_006103_050 |
0.1367343406 |
- |
- |
- |
| 15 |
Hb_020112_010 |
0.1399946348 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 16 |
Hb_135673_010 |
0.1418110473 |
- |
- |
Germin-like protein subfamily 1 member 6 precursor [Populus trichocarpa] |
| 17 |
Hb_151094_020 |
0.1422704174 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Prunus mume] |
| 18 |
Hb_000732_200 |
0.1437604677 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000984_280 |
0.1460811106 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 2 [Jatropha curcas] |
| 20 |
Hb_004330_050 |
0.1487591498 |
- |
- |
conserved hypothetical protein [Ricinus communis] |