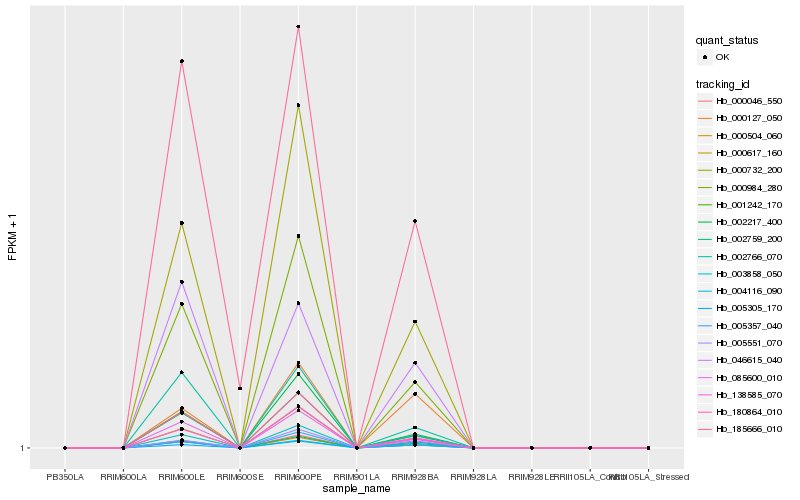
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002759_200 |
0.0 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 2 [Jatropha curcas] |
| 2 |
Hb_000504_060 |
0.0406064181 |
- |
- |
Cell division cycle protein cdt2, putative [Ricinus communis] |
| 3 |
Hb_005305_170 |
0.0652840277 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 8 [Jatropha curcas] |
| 4 |
Hb_003858_050 |
0.0693012641 |
- |
- |
hypothetical protein PRUPE_ppa021882mg [Prunus persica] |
| 5 |
Hb_000046_550 |
0.0701119768 |
- |
- |
PREDICTED: protein ALWAYS EARLY 3 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000984_280 |
0.0827962631 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 2 [Jatropha curcas] |
| 7 |
Hb_005551_070 |
0.0854231203 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000732_200 |
0.0909187762 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_085600_010 |
0.0938443382 |
- |
- |
PREDICTED: peroxidase 24-like [Jatropha curcas] |
| 10 |
Hb_001242_170 |
0.0980607513 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000617_160 |
0.1018493083 |
- |
- |
- |
| 12 |
Hb_138585_070 |
0.1056614687 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004116_090 |
0.1105095836 |
- |
- |
- |
| 14 |
Hb_046615_040 |
0.1199946802 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_180864_010 |
0.1212181975 |
- |
- |
hypothetical protein B456_010G097000 [Gossypium raimondii] |
| 16 |
Hb_000127_050 |
0.1252791463 |
- |
- |
- |
| 17 |
Hb_005357_040 |
0.1273205634 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 18 |
Hb_002766_070 |
0.127371391 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_17894 [Jatropha curcas] |
| 19 |
Hb_002217_400 |
0.1296666702 |
- |
- |
polynucleotidyl transferase, ribonuclease H-like protein [Arabidopsis thaliana] |
| 20 |
Hb_185666_010 |
0.1333941496 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3 [Jatropha curcas] |