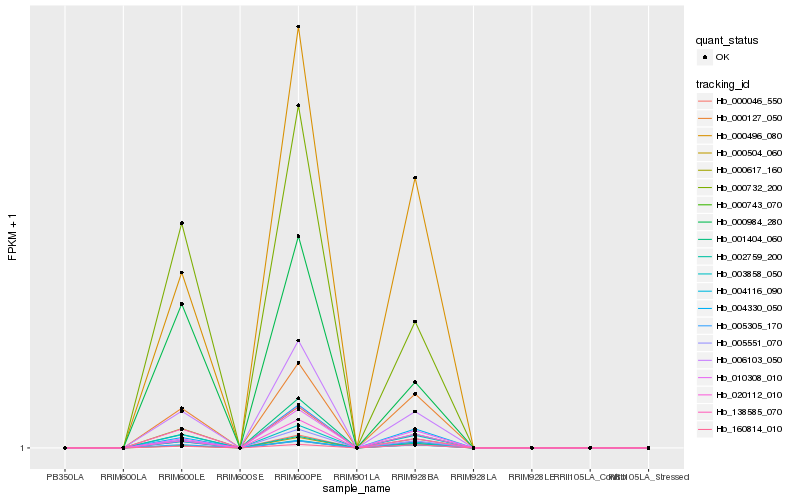
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000046_550 |
0.0 |
- |
- |
PREDICTED: protein ALWAYS EARLY 3 isoform X2 [Jatropha curcas] |
| 2 |
Hb_005305_170 |
0.0137021752 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 8 [Jatropha curcas] |
| 3 |
Hb_003858_050 |
0.031950251 |
- |
- |
hypothetical protein PRUPE_ppa021882mg [Prunus persica] |
| 4 |
Hb_005551_070 |
0.0461790857 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000127_050 |
0.0572831265 |
- |
- |
- |
| 6 |
Hb_002759_200 |
0.0701119768 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 2 [Jatropha curcas] |
| 7 |
Hb_000743_070 |
0.1003380872 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21060 [Jatropha curcas] |
| 8 |
Hb_000504_060 |
0.1040575816 |
- |
- |
Cell division cycle protein cdt2, putative [Ricinus communis] |
| 9 |
Hb_004116_090 |
0.1048180268 |
- |
- |
- |
| 10 |
Hb_006103_050 |
0.1049765059 |
- |
- |
- |
| 11 |
Hb_020112_010 |
0.1173524842 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 12 |
Hb_004330_050 |
0.1229759689 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000732_200 |
0.1250057736 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000496_080 |
0.1256500005 |
- |
- |
Proteinase inhibitor, putative [Ricinus communis] |
| 15 |
Hb_010308_010 |
0.1261794037 |
- |
- |
hypothetical protein JCGZ_22818 [Jatropha curcas] |
| 16 |
Hb_000617_160 |
0.1280826691 |
- |
- |
- |
| 17 |
Hb_138585_070 |
0.1281962257 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001404_060 |
0.1319603111 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_160814_010 |
0.1321749055 |
- |
- |
PREDICTED: probable sulfate transporter 3.5 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000984_280 |
0.1339102568 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 2 [Jatropha curcas] |