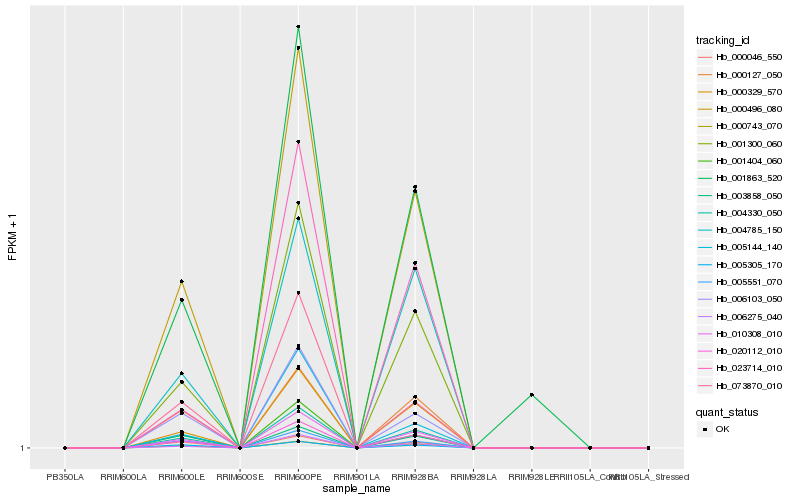
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000496_080 |
0.0 |
- |
- |
Proteinase inhibitor, putative [Ricinus communis] |
| 2 |
Hb_010308_010 |
0.0011774589 |
- |
- |
hypothetical protein JCGZ_22818 [Jatropha curcas] |
| 3 |
Hb_004330_050 |
0.0063438843 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_020112_010 |
0.0176387648 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 5 |
Hb_001300_060 |
0.0342165881 |
- |
- |
PREDICTED: monothiol glutaredoxin-S6-like [Jatropha curcas] |
| 6 |
Hb_000329_570 |
0.0534028346 |
- |
- |
- |
| 7 |
Hb_000743_070 |
0.0709936341 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21060 [Jatropha curcas] |
| 8 |
Hb_006103_050 |
0.0902254579 |
- |
- |
- |
| 9 |
Hb_004785_150 |
0.0908276412 |
- |
- |
hypothetical protein JCGZ_25207 [Jatropha curcas] |
| 10 |
Hb_006275_040 |
0.0914239498 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 11 |
Hb_001404_060 |
0.0971049116 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_000127_050 |
0.1021700176 |
- |
- |
- |
| 13 |
Hb_005551_070 |
0.1057860764 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 14 |
Hb_073870_010 |
0.109431014 |
- |
- |
hypothetical protein RCOM_0725510 [Ricinus communis] |
| 15 |
Hb_005305_170 |
0.1246541652 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 8 [Jatropha curcas] |
| 16 |
Hb_000046_550 |
0.1256500005 |
- |
- |
PREDICTED: protein ALWAYS EARLY 3 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001863_520 |
0.128656917 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_023714_010 |
0.1404645284 |
- |
- |
hypothetical protein B456_007G377100 [Gossypium raimondii] |
| 19 |
Hb_005144_140 |
0.1408951326 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003858_050 |
0.1505957318 |
- |
- |
hypothetical protein PRUPE_ppa021882mg [Prunus persica] |