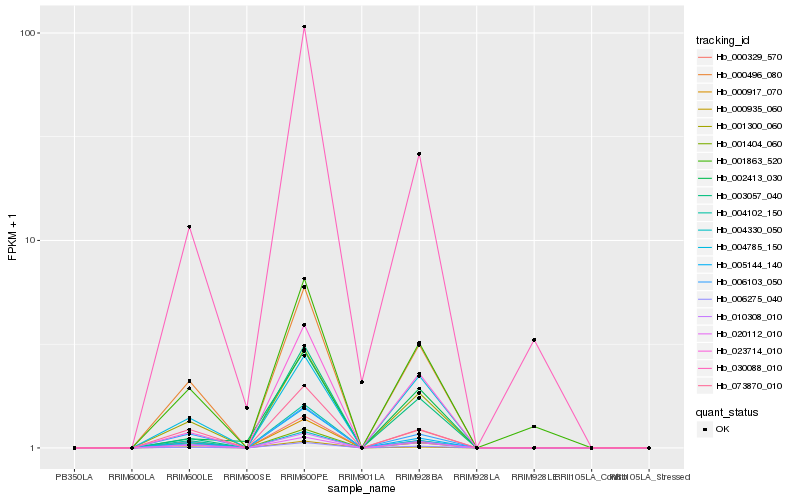
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006275_040 |
0.0 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 2 |
Hb_001300_060 |
0.0645210781 |
- |
- |
PREDICTED: monothiol glutaredoxin-S6-like [Jatropha curcas] |
| 3 |
Hb_005144_140 |
0.0708703284 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003057_040 |
0.0818757464 |
- |
- |
PREDICTED: salt stress-induced hydrophobic peptide ESI3 [Vitis vinifera] |
| 5 |
Hb_000917_070 |
0.0845473381 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_010308_010 |
0.0904369257 |
- |
- |
hypothetical protein JCGZ_22818 [Jatropha curcas] |
| 7 |
Hb_023714_010 |
0.0905758736 |
- |
- |
hypothetical protein B456_007G377100 [Gossypium raimondii] |
| 8 |
Hb_004330_050 |
0.0911795511 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000496_080 |
0.0914239498 |
- |
- |
Proteinase inhibitor, putative [Ricinus communis] |
| 10 |
Hb_000329_570 |
0.0967211351 |
- |
- |
- |
| 11 |
Hb_020112_010 |
0.1073251685 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 12 |
Hb_073870_010 |
0.1131904839 |
- |
- |
hypothetical protein RCOM_0725510 [Ricinus communis] |
| 13 |
Hb_001404_060 |
0.1181488075 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 14 |
Hb_030088_010 |
0.1271636737 |
- |
- |
basic blue copper family protein [Populus trichocarpa] |
| 15 |
Hb_004102_150 |
0.1299126621 |
- |
- |
Late embryogenesis abundant protein D-7, putative [Ricinus communis] |
| 16 |
Hb_006103_050 |
0.1321566005 |
- |
- |
- |
| 17 |
Hb_001863_520 |
0.1347953513 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000935_060 |
0.1381559185 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 8-like [Jatropha curcas] |
| 19 |
Hb_004785_150 |
0.1520432302 |
- |
- |
hypothetical protein JCGZ_25207 [Jatropha curcas] |
| 20 |
Hb_002413_030 |
0.1577531429 |
- |
- |
sulfate transporter, putative [Ricinus communis] |