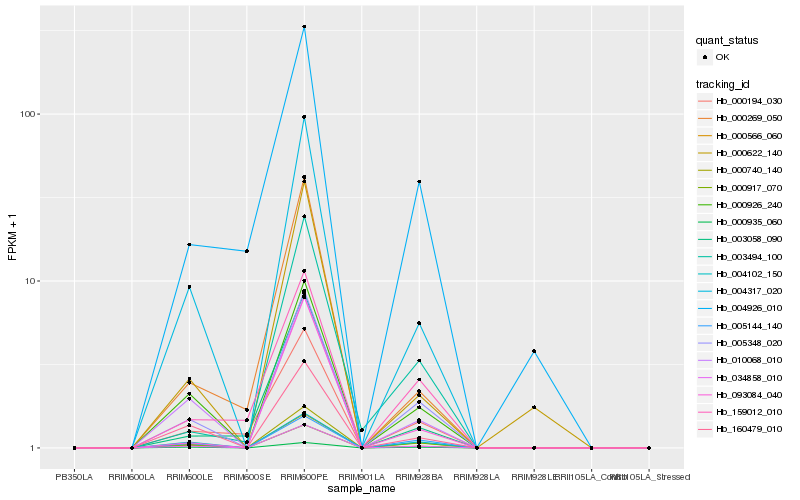
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000740_140 |
0.0 |
transcription factor |
TF Family: OFP |
PREDICTED: probable transcription repressor OFP9 [Jatropha curcas] |
| 2 |
Hb_005348_020 |
0.0237079209 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_034858_010 |
0.0537071284 |
- |
- |
- |
| 4 |
Hb_004317_020 |
0.0895976639 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7-like [Malus domestica] |
| 5 |
Hb_000926_240 |
0.0939180175 |
- |
- |
hypothetical protein PRUPE_ppb018041mg [Prunus persica] |
| 6 |
Hb_000566_060 |
0.1030481856 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000917_070 |
0.104114017 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004102_150 |
0.1075425698 |
- |
- |
Late embryogenesis abundant protein D-7, putative [Ricinus communis] |
| 9 |
Hb_010068_010 |
0.1155854391 |
- |
- |
PREDICTED: uncharacterized protein LOC105633273 [Jatropha curcas] |
| 10 |
Hb_000935_060 |
0.1164389575 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 8-like [Jatropha curcas] |
| 11 |
Hb_005144_140 |
0.1210168228 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003494_100 |
0.1274885691 |
transcription factor |
TF Family: zf-HD |
Mini zinc finger 2 isoform 1 [Theobroma cacao] |
| 13 |
Hb_093084_040 |
0.1286577783 |
transcription factor |
TF Family: LOB |
hypothetical protein POPTR_0015s09370g [Populus trichocarpa] |
| 14 |
Hb_160479_010 |
0.135142097 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 15 |
Hb_000269_050 |
0.1442113918 |
- |
- |
PREDICTED: beta-glucosidase 12-like [Gossypium raimondii] |
| 16 |
Hb_003058_090 |
0.1442654099 |
- |
- |
hypothetical protein POPTR_0001s28080g [Populus trichocarpa] |
| 17 |
Hb_000194_030 |
0.1450224768 |
transcription factor |
TF Family: NAC |
NAC transcription factor 059 [Jatropha curcas] |
| 18 |
Hb_159012_010 |
0.1458030203 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g06940 [Jatropha curcas] |
| 19 |
Hb_004926_010 |
0.1473171002 |
- |
- |
hypothetical protein JCGZ_27059 [Jatropha curcas] |
| 20 |
Hb_000622_140 |
0.1482936977 |
- |
- |
conserved hypothetical protein [Ricinus communis] |