| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
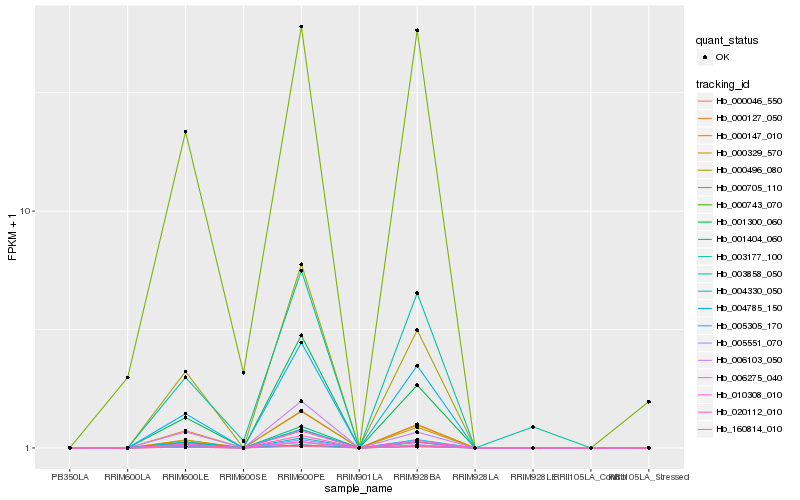
Hb_000743_070 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21060 [Jatropha curcas] |
| 2 |
Hb_000127_050 |
0.0496997134 |
- |
- |
- |
| 3 |
Hb_020112_010 |
0.0534752934 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 4 |
Hb_004785_150 |
0.0637897835 |
- |
- |
hypothetical protein JCGZ_25207 [Jatropha curcas] |
| 5 |
Hb_000496_080 |
0.0709936341 |
- |
- |
Proteinase inhibitor, putative [Ricinus communis] |
| 6 |
Hb_010308_010 |
0.0721499842 |
- |
- |
hypothetical protein JCGZ_22818 [Jatropha curcas] |
| 7 |
Hb_004330_050 |
0.0740013913 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000329_570 |
0.0799444884 |
- |
- |
- |
| 9 |
Hb_160814_010 |
0.0904682761 |
- |
- |
PREDICTED: probable sulfate transporter 3.5 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001300_060 |
0.0955354647 |
- |
- |
PREDICTED: monothiol glutaredoxin-S6-like [Jatropha curcas] |
| 11 |
Hb_000046_550 |
0.1003380872 |
- |
- |
PREDICTED: protein ALWAYS EARLY 3 isoform X2 [Jatropha curcas] |
| 12 |
Hb_005305_170 |
0.1072296348 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 8 [Jatropha curcas] |
| 13 |
Hb_005551_070 |
0.1094112498 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003858_050 |
0.1120827798 |
- |
- |
hypothetical protein PRUPE_ppa021882mg [Prunus persica] |
| 15 |
Hb_006103_050 |
0.1306392905 |
- |
- |
- |
| 16 |
Hb_000705_110 |
0.1332615024 |
- |
- |
hypothetical protein POPTR_0005s09220g [Populus trichocarpa] |
| 17 |
Hb_000147_010 |
0.1483070362 |
- |
- |
PREDICTED: UDP-glycosyltransferase 87A1-like [Jatropha curcas] |
| 18 |
Hb_001404_060 |
0.1486019149 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_003177_100 |
0.1506581414 |
- |
- |
PREDICTED: auxin-induced protein 6B [Jatropha curcas] |
| 20 |
Hb_006275_040 |
0.159000625 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |