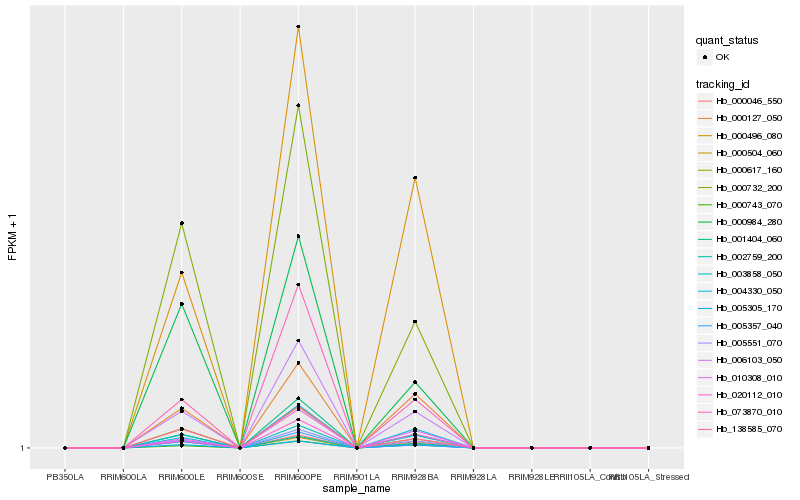
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005551_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005305_170 |
0.0347387766 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 8 [Jatropha curcas] |
| 3 |
Hb_000046_550 |
0.0461790857 |
- |
- |
PREDICTED: protein ALWAYS EARLY 3 isoform X2 [Jatropha curcas] |
| 4 |
Hb_006103_050 |
0.059993121 |
- |
- |
- |
| 5 |
Hb_003858_050 |
0.0776570498 |
- |
- |
hypothetical protein PRUPE_ppa021882mg [Prunus persica] |
| 6 |
Hb_000127_050 |
0.0835355333 |
- |
- |
- |
| 7 |
Hb_002759_200 |
0.0854231203 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 2 [Jatropha curcas] |
| 8 |
Hb_001404_060 |
0.0870518568 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_138585_070 |
0.0956843314 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000617_160 |
0.097611237 |
- |
- |
- |
| 11 |
Hb_000732_200 |
0.0995914426 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004330_050 |
0.1012373823 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_020112_010 |
0.1038511684 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 14 |
Hb_000504_060 |
0.1042579852 |
- |
- |
Cell division cycle protein cdt2, putative [Ricinus communis] |
| 15 |
Hb_000496_080 |
0.1057860764 |
- |
- |
Proteinase inhibitor, putative [Ricinus communis] |
| 16 |
Hb_010308_010 |
0.1059127139 |
- |
- |
hypothetical protein JCGZ_22818 [Jatropha curcas] |
| 17 |
Hb_005357_040 |
0.1076751224 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 18 |
Hb_073870_010 |
0.1082244081 |
- |
- |
hypothetical protein RCOM_0725510 [Ricinus communis] |
| 19 |
Hb_000743_070 |
0.1094112498 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21060 [Jatropha curcas] |
| 20 |
Hb_000984_280 |
0.1188028434 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 2 [Jatropha curcas] |