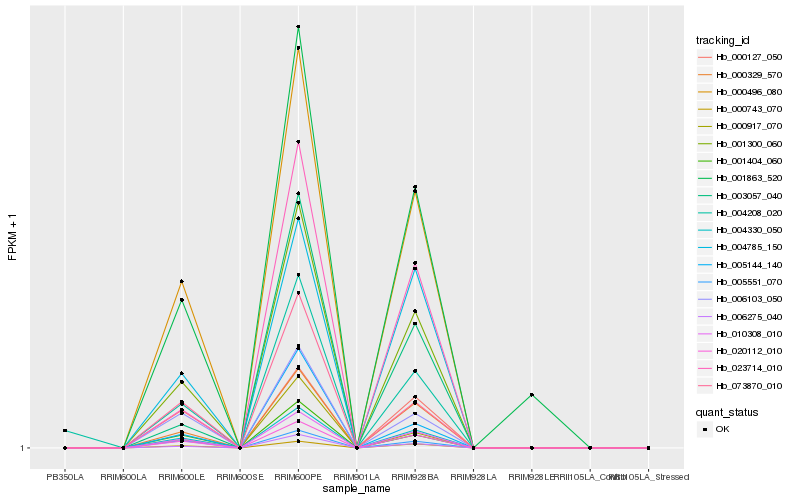
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001300_060 |
0.0 |
- |
- |
PREDICTED: monothiol glutaredoxin-S6-like [Jatropha curcas] |
| 2 |
Hb_010308_010 |
0.0337028968 |
- |
- |
hypothetical protein JCGZ_22818 [Jatropha curcas] |
| 3 |
Hb_000496_080 |
0.0342165881 |
- |
- |
Proteinase inhibitor, putative [Ricinus communis] |
| 4 |
Hb_004330_050 |
0.0374911395 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000329_570 |
0.03995332 |
- |
- |
- |
| 6 |
Hb_020112_010 |
0.0460393135 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 7 |
Hb_006275_040 |
0.0645210781 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 8 |
Hb_004785_150 |
0.0929052247 |
- |
- |
hypothetical protein JCGZ_25207 [Jatropha curcas] |
| 9 |
Hb_000743_070 |
0.0955354647 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21060 [Jatropha curcas] |
| 10 |
Hb_023714_010 |
0.107573514 |
- |
- |
hypothetical protein B456_007G377100 [Gossypium raimondii] |
| 11 |
Hb_001404_060 |
0.112669255 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_006103_050 |
0.1130751865 |
- |
- |
- |
| 13 |
Hb_073870_010 |
0.1194700787 |
- |
- |
hypothetical protein RCOM_0725510 [Ricinus communis] |
| 14 |
Hb_003057_040 |
0.1232190517 |
- |
- |
PREDICTED: salt stress-induced hydrophobic peptide ESI3 [Vitis vinifera] |
| 15 |
Hb_001863_520 |
0.1235241683 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005144_140 |
0.1252954129 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000127_050 |
0.133294557 |
- |
- |
- |
| 18 |
Hb_005551_070 |
0.1378445439 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000917_070 |
0.1413685288 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004208_020 |
0.1523552033 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |