| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
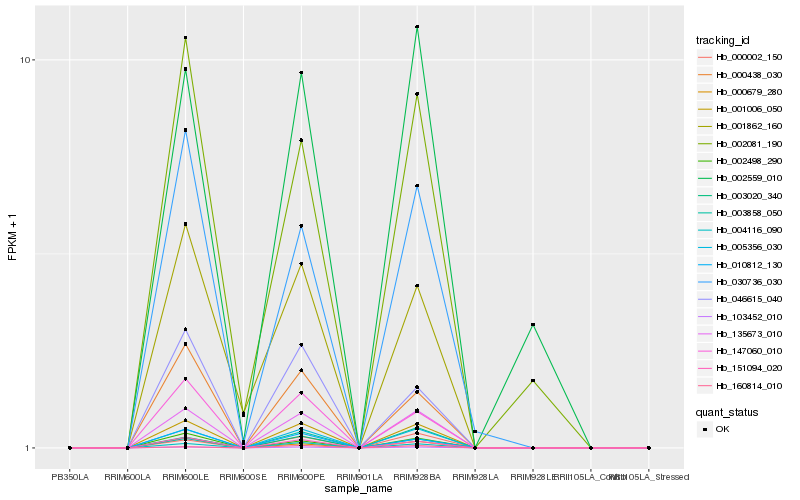
Hb_135673_010 |
0.0 |
- |
- |
Germin-like protein subfamily 1 member 6 precursor [Populus trichocarpa] |
| 2 |
Hb_103452_010 |
0.0145875702 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 3 |
Hb_001006_050 |
0.0194031408 |
- |
- |
PREDICTED: uncharacterized protein LOC101777526 [Setaria italica] |
| 4 |
Hb_005356_030 |
0.0253422989 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_004116_090 |
0.0693937684 |
- |
- |
- |
| 6 |
Hb_151094_020 |
0.0697537454 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Prunus mume] |
| 7 |
Hb_002498_290 |
0.1101366163 |
- |
- |
PREDICTED: autophagy-related protein 8C-like isoform X2 [Elaeis guineensis] |
| 8 |
Hb_000002_150 |
0.1192324066 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_147060_010 |
0.1199211788 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Populus euphratica] |
| 10 |
Hb_000438_030 |
0.1214765208 |
- |
- |
- |
| 11 |
Hb_010812_130 |
0.121672594 |
- |
- |
PREDICTED: hydroxyacylglutathione hydrolase cytoplasmic [Jatropha curcas] |
| 12 |
Hb_030736_030 |
0.1244818099 |
- |
- |
hypothetical protein VITISV_006651 [Vitis vinifera] |
| 13 |
Hb_000679_280 |
0.1300829195 |
- |
- |
hypothetical protein POPTR_0006s27120g [Populus trichocarpa] |
| 14 |
Hb_003020_340 |
0.1311315287 |
- |
- |
PREDICTED: uncharacterized protein LOC105631327 [Jatropha curcas] |
| 15 |
Hb_046615_040 |
0.1413761996 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_003858_050 |
0.1418110473 |
- |
- |
hypothetical protein PRUPE_ppa021882mg [Prunus persica] |
| 17 |
Hb_160814_010 |
0.1503138662 |
- |
- |
PREDICTED: probable sulfate transporter 3.5 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002559_010 |
0.1522289 |
- |
- |
hypothetical protein JCGZ_06274 [Jatropha curcas] |
| 19 |
Hb_001862_160 |
0.1555738296 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.1 [Jatropha curcas] |
| 20 |
Hb_002081_190 |
0.1560796352 |
- |
- |
hypothetical protein EUGRSUZ_I010391, partial [Eucalyptus grandis] |