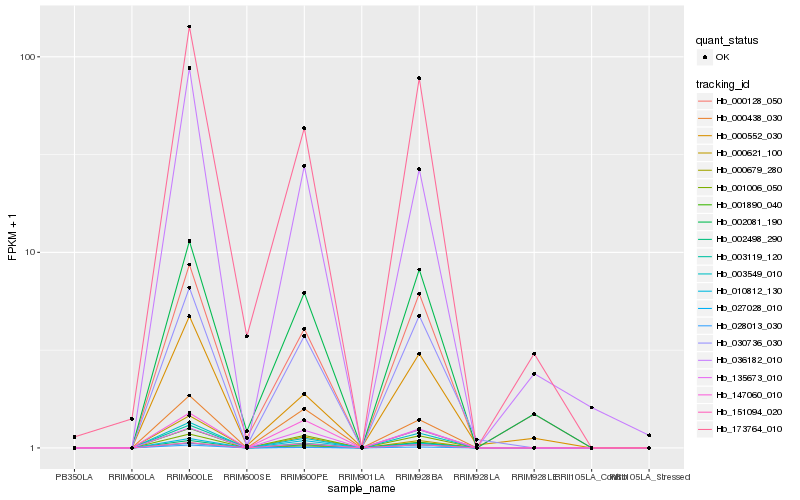
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003549_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647434 [Jatropha curcas] |
| 2 |
Hb_000679_280 |
0.037339964 |
- |
- |
hypothetical protein POPTR_0006s27120g [Populus trichocarpa] |
| 3 |
Hb_002498_290 |
0.0528799949 |
- |
- |
PREDICTED: autophagy-related protein 8C-like isoform X2 [Elaeis guineensis] |
| 4 |
Hb_030736_030 |
0.0955508285 |
- |
- |
hypothetical protein VITISV_006651 [Vitis vinifera] |
| 5 |
Hb_173764_010 |
0.1024496372 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_027028_010 |
0.1129977554 |
- |
- |
hypothetical protein JCGZ_01141 [Jatropha curcas] |
| 7 |
Hb_003119_120 |
0.1255974701 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Nicotiana sylvestris] |
| 8 |
Hb_000552_030 |
0.1320398752 |
- |
- |
glucose-6-phosphate 1-dehydrogenase, putative [Ricinus communis] |
| 9 |
Hb_028013_030 |
0.1354854676 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_036182_010 |
0.1364751288 |
- |
- |
hypothetical protein POPTR_0473s00220g [Populus trichocarpa] |
| 11 |
Hb_000438_030 |
0.1388871967 |
- |
- |
- |
| 12 |
Hb_002081_190 |
0.1400274228 |
- |
- |
hypothetical protein EUGRSUZ_I010391, partial [Eucalyptus grandis] |
| 13 |
Hb_000128_050 |
0.1436293955 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_151094_020 |
0.1460696755 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Prunus mume] |
| 15 |
Hb_001890_040 |
0.1497863503 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 16 |
Hb_147060_010 |
0.1576415906 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Populus euphratica] |
| 17 |
Hb_010812_130 |
0.1608292381 |
- |
- |
PREDICTED: hydroxyacylglutathione hydrolase cytoplasmic [Jatropha curcas] |
| 18 |
Hb_135673_010 |
0.1623892515 |
- |
- |
Germin-like protein subfamily 1 member 6 precursor [Populus trichocarpa] |
| 19 |
Hb_001006_050 |
0.1634108944 |
- |
- |
PREDICTED: uncharacterized protein LOC101777526 [Setaria italica] |
| 20 |
Hb_000621_100 |
0.1651215412 |
- |
- |
conserved hypothetical protein [Ricinus communis] |