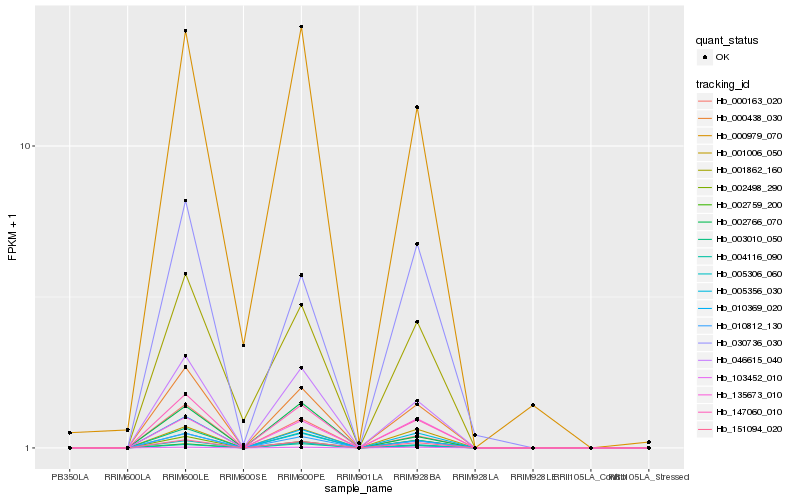
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_147060_010 |
0.0 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Populus euphratica] |
| 2 |
Hb_010812_130 |
0.0032995124 |
- |
- |
PREDICTED: hydroxyacylglutathione hydrolase cytoplasmic [Jatropha curcas] |
| 3 |
Hb_000438_030 |
0.0202660909 |
- |
- |
- |
| 4 |
Hb_046615_040 |
0.0284786124 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_003010_050 |
0.039787954 |
- |
- |
- |
| 6 |
Hb_151094_020 |
0.0504873264 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Prunus mume] |
| 7 |
Hb_005306_060 |
0.0674046815 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 5 [Jatropha curcas] |
| 8 |
Hb_000163_020 |
0.0989843068 |
- |
- |
- |
| 9 |
Hb_001006_050 |
0.1025447622 |
- |
- |
PREDICTED: uncharacterized protein LOC101777526 [Setaria italica] |
| 10 |
Hb_004116_090 |
0.1170128374 |
- |
- |
- |
| 11 |
Hb_103452_010 |
0.1198230272 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 12 |
Hb_135673_010 |
0.1199211788 |
- |
- |
Germin-like protein subfamily 1 member 6 precursor [Populus trichocarpa] |
| 13 |
Hb_010369_020 |
0.1209464408 |
- |
- |
hypothetical protein JCGZ_21088 [Jatropha curcas] |
| 14 |
Hb_002498_290 |
0.1291178614 |
- |
- |
PREDICTED: autophagy-related protein 8C-like isoform X2 [Elaeis guineensis] |
| 15 |
Hb_005356_030 |
0.1310126948 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 16 |
Hb_002759_200 |
0.1361894162 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 2 [Jatropha curcas] |
| 17 |
Hb_000979_070 |
0.1380005542 |
- |
- |
PREDICTED: endoglucanase 8-like [Jatropha curcas] |
| 18 |
Hb_001862_160 |
0.1392083202 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.1 [Jatropha curcas] |
| 19 |
Hb_030736_030 |
0.1431848397 |
- |
- |
hypothetical protein VITISV_006651 [Vitis vinifera] |
| 20 |
Hb_002766_070 |
0.1463823572 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_17894 [Jatropha curcas] |