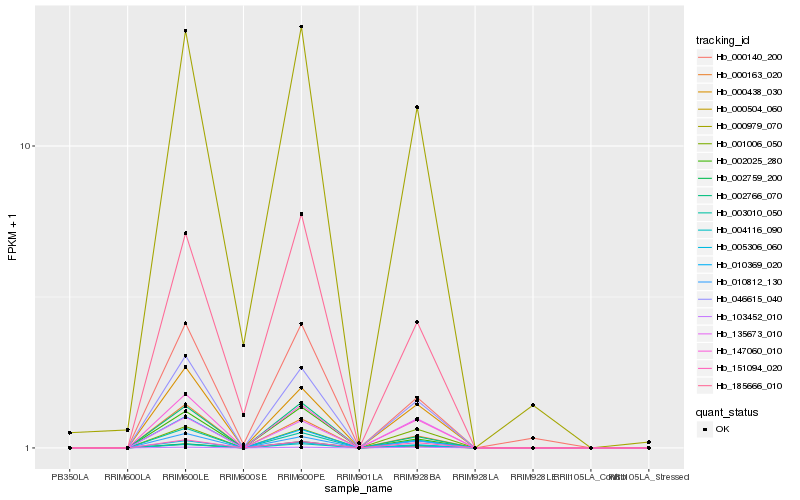
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_046615_040 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_003010_050 |
0.0217381571 |
- |
- |
- |
| 3 |
Hb_010812_130 |
0.0252854554 |
- |
- |
PREDICTED: hydroxyacylglutathione hydrolase cytoplasmic [Jatropha curcas] |
| 4 |
Hb_147060_010 |
0.0284786124 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Populus euphratica] |
| 5 |
Hb_000438_030 |
0.0451345982 |
- |
- |
- |
| 6 |
Hb_005306_060 |
0.0471132509 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 5 [Jatropha curcas] |
| 7 |
Hb_151094_020 |
0.0741110258 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Prunus mume] |
| 8 |
Hb_000163_020 |
0.086322596 |
- |
- |
- |
| 9 |
Hb_010369_020 |
0.1114115661 |
- |
- |
hypothetical protein JCGZ_21088 [Jatropha curcas] |
| 10 |
Hb_002766_070 |
0.1182877989 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_17894 [Jatropha curcas] |
| 11 |
Hb_002759_200 |
0.1199946802 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 2 [Jatropha curcas] |
| 12 |
Hb_001006_050 |
0.1229529606 |
- |
- |
PREDICTED: uncharacterized protein LOC101777526 [Setaria italica] |
| 13 |
Hb_000504_060 |
0.125417647 |
- |
- |
Cell division cycle protein cdt2, putative [Ricinus communis] |
| 14 |
Hb_004116_090 |
0.1256225907 |
- |
- |
- |
| 15 |
Hb_002025_280 |
0.1333320725 |
- |
- |
PREDICTED: uncharacterized protein LOC105634201 [Jatropha curcas] |
| 16 |
Hb_000979_070 |
0.1342860164 |
- |
- |
PREDICTED: endoglucanase 8-like [Jatropha curcas] |
| 17 |
Hb_103452_010 |
0.1390923404 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 18 |
Hb_185666_010 |
0.141178752 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3 [Jatropha curcas] |
| 19 |
Hb_135673_010 |
0.1413761996 |
- |
- |
Germin-like protein subfamily 1 member 6 precursor [Populus trichocarpa] |
| 20 |
Hb_000140_200 |
0.1421533675 |
- |
- |
PREDICTED: uncharacterized protein LOC105642928 [Jatropha curcas] |