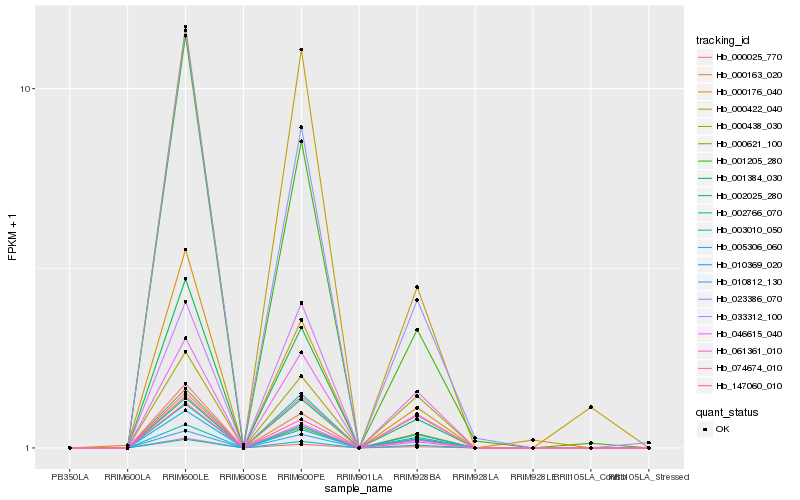
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000163_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_010369_020 |
0.0267914365 |
- |
- |
hypothetical protein JCGZ_21088 [Jatropha curcas] |
| 3 |
Hb_005306_060 |
0.0431131957 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 5 [Jatropha curcas] |
| 4 |
Hb_003010_050 |
0.0647345031 |
- |
- |
- |
| 5 |
Hb_046615_040 |
0.086322596 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_074674_010 |
0.08843975 |
- |
- |
Cell division cycle protein cdt2, putative [Ricinus communis] |
| 7 |
Hb_000438_030 |
0.0972311444 |
- |
- |
- |
| 8 |
Hb_010812_130 |
0.0978854554 |
- |
- |
PREDICTED: hydroxyacylglutathione hydrolase cytoplasmic [Jatropha curcas] |
| 9 |
Hb_147060_010 |
0.0989843068 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Populus euphratica] |
| 10 |
Hb_001384_030 |
0.1013992866 |
- |
- |
PREDICTED: uncharacterized protein LOC105049010 [Elaeis guineensis] |
| 11 |
Hb_023386_070 |
0.1066207583 |
- |
- |
PREDICTED: probable pectinesterase 68 [Jatropha curcas] |
| 12 |
Hb_061361_010 |
0.1150737577 |
- |
- |
kinase, putative [Ricinus communis] |
| 13 |
Hb_002766_070 |
0.1161122906 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_17894 [Jatropha curcas] |
| 14 |
Hb_000025_770 |
0.1198451069 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 15 |
Hb_033312_100 |
0.1245164595 |
- |
- |
- |
| 16 |
Hb_002025_280 |
0.1273878763 |
- |
- |
PREDICTED: uncharacterized protein LOC105634201 [Jatropha curcas] |
| 17 |
Hb_001205_280 |
0.1308504005 |
- |
- |
PREDICTED: probable polyol transporter 6 [Jatropha curcas] |
| 18 |
Hb_000176_040 |
0.1314445986 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHR3 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000422_040 |
0.1330655101 |
- |
- |
hypothetical protein CICLE_v10013147mg [Citrus clementina] |
| 20 |
Hb_000621_100 |
0.1336107768 |
- |
- |
conserved hypothetical protein [Ricinus communis] |