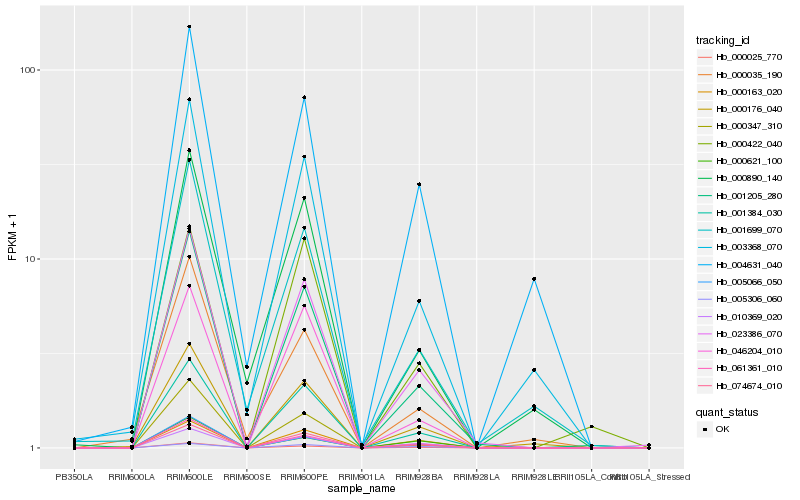
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023386_070 |
0.0 |
- |
- |
PREDICTED: probable pectinesterase 68 [Jatropha curcas] |
| 2 |
Hb_001205_280 |
0.0546757986 |
- |
- |
PREDICTED: probable polyol transporter 6 [Jatropha curcas] |
| 3 |
Hb_074674_010 |
0.0625795327 |
- |
- |
Cell division cycle protein cdt2, putative [Ricinus communis] |
| 4 |
Hb_001384_030 |
0.0643670761 |
- |
- |
PREDICTED: uncharacterized protein LOC105049010 [Elaeis guineensis] |
| 5 |
Hb_061361_010 |
0.0721124319 |
- |
- |
kinase, putative [Ricinus communis] |
| 6 |
Hb_000025_770 |
0.076070124 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 7 |
Hb_010369_020 |
0.0886698923 |
- |
- |
hypothetical protein JCGZ_21088 [Jatropha curcas] |
| 8 |
Hb_000176_040 |
0.1043988211 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHR3 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000163_020 |
0.1066207583 |
- |
- |
- |
| 10 |
Hb_005066_050 |
0.1100465636 |
- |
- |
- |
| 11 |
Hb_046204_010 |
0.1176098162 |
- |
- |
PREDICTED: peroxidase 5-like [Jatropha curcas] |
| 12 |
Hb_003368_070 |
0.1189153747 |
- |
- |
PREDICTED: uncharacterized protein LOC105109387 [Populus euphratica] |
| 13 |
Hb_000347_310 |
0.1286882049 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 3-like [Jatropha curcas] |
| 14 |
Hb_001699_070 |
0.131862989 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000621_100 |
0.1358628769 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000422_040 |
0.1387145572 |
- |
- |
hypothetical protein CICLE_v10013147mg [Citrus clementina] |
| 17 |
Hb_004631_040 |
0.1387957218 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 18 |
Hb_000890_140 |
0.139246188 |
- |
- |
PREDICTED: uncharacterized protein LOC105646185 [Jatropha curcas] |
| 19 |
Hb_000035_190 |
0.1416833179 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005306_060 |
0.1425029934 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 5 [Jatropha curcas] |