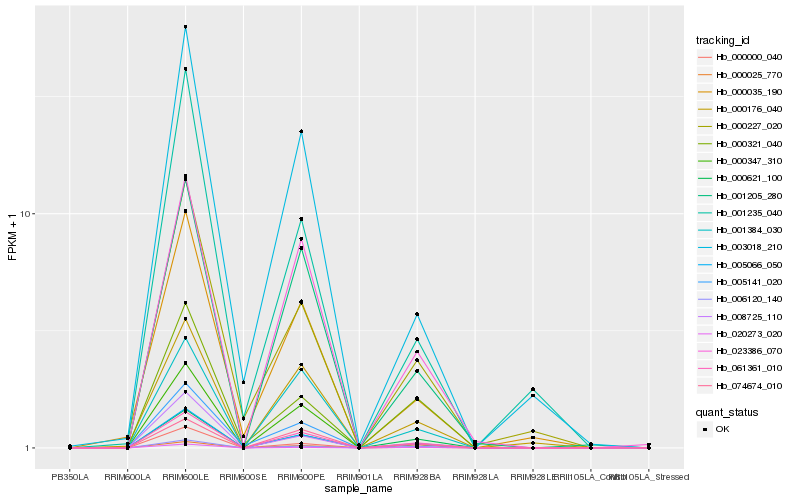
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005066_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_000025_770 |
0.0453179799 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 3 |
Hb_061361_010 |
0.0509348032 |
- |
- |
kinase, putative [Ricinus communis] |
| 4 |
Hb_000000_040 |
0.0773566774 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075-like [Jatropha curcas] |
| 5 |
Hb_000621_100 |
0.0887405243 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_008725_110 |
0.0951696538 |
- |
- |
PREDICTED: leucine-rich repeat extensin-like protein 1 [Jatropha curcas] |
| 7 |
Hb_001205_280 |
0.0979399288 |
- |
- |
PREDICTED: probable polyol transporter 6 [Jatropha curcas] |
| 8 |
Hb_006120_140 |
0.1014078054 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 9 |
Hb_023386_070 |
0.1100465636 |
- |
- |
PREDICTED: probable pectinesterase 68 [Jatropha curcas] |
| 10 |
Hb_005141_020 |
0.1205763387 |
- |
- |
laccase, putative [Ricinus communis] |
| 11 |
Hb_000347_310 |
0.1208114429 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 3-like [Jatropha curcas] |
| 12 |
Hb_020273_020 |
0.1233356636 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 13 |
Hb_000035_190 |
0.1237990196 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000227_020 |
0.1275527919 |
- |
- |
PREDICTED: basic 7S globulin-like [Jatropha curcas] |
| 15 |
Hb_003018_210 |
0.127867583 |
- |
- |
PREDICTED: uncharacterized protein LOC105138721 isoform X2 [Populus euphratica] |
| 16 |
Hb_000176_040 |
0.1302897731 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHR3 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000321_040 |
0.1303607832 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 5 [Jatropha curcas] |
| 18 |
Hb_001384_030 |
0.1306286765 |
- |
- |
PREDICTED: uncharacterized protein LOC105049010 [Elaeis guineensis] |
| 19 |
Hb_074674_010 |
0.1309526669 |
- |
- |
Cell division cycle protein cdt2, putative [Ricinus communis] |
| 20 |
Hb_001235_040 |
0.1345276758 |
- |
- |
hypothetical protein POPTR_0001s47470g [Populus trichocarpa] |