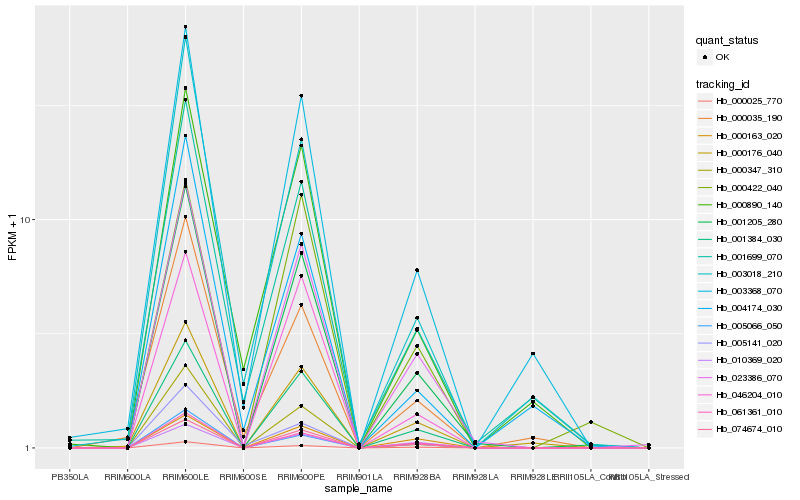
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001205_280 |
0.0 |
- |
- |
PREDICTED: probable polyol transporter 6 [Jatropha curcas] |
| 2 |
Hb_023386_070 |
0.0546757986 |
- |
- |
PREDICTED: probable pectinesterase 68 [Jatropha curcas] |
| 3 |
Hb_001384_030 |
0.0637789003 |
- |
- |
PREDICTED: uncharacterized protein LOC105049010 [Elaeis guineensis] |
| 4 |
Hb_074674_010 |
0.0678969753 |
- |
- |
Cell division cycle protein cdt2, putative [Ricinus communis] |
| 5 |
Hb_061361_010 |
0.0689216143 |
- |
- |
kinase, putative [Ricinus communis] |
| 6 |
Hb_000025_770 |
0.071918301 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 7 |
Hb_000347_310 |
0.0978307675 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 3-like [Jatropha curcas] |
| 8 |
Hb_005066_050 |
0.0979399288 |
- |
- |
- |
| 9 |
Hb_000176_040 |
0.1042122688 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHR3 isoform X2 [Jatropha curcas] |
| 10 |
Hb_046204_010 |
0.1105907546 |
- |
- |
PREDICTED: peroxidase 5-like [Jatropha curcas] |
| 11 |
Hb_010369_020 |
0.1108609737 |
- |
- |
hypothetical protein JCGZ_21088 [Jatropha curcas] |
| 12 |
Hb_003368_070 |
0.1153587331 |
- |
- |
PREDICTED: uncharacterized protein LOC105109387 [Populus euphratica] |
| 13 |
Hb_001699_070 |
0.1256933518 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003018_210 |
0.1305028648 |
- |
- |
PREDICTED: uncharacterized protein LOC105138721 isoform X2 [Populus euphratica] |
| 15 |
Hb_000163_020 |
0.1308504005 |
- |
- |
- |
| 16 |
Hb_000035_190 |
0.1345102254 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000422_040 |
0.1350228195 |
- |
- |
hypothetical protein CICLE_v10013147mg [Citrus clementina] |
| 18 |
Hb_005141_020 |
0.1351547341 |
- |
- |
laccase, putative [Ricinus communis] |
| 19 |
Hb_000890_140 |
0.1383810976 |
- |
- |
PREDICTED: uncharacterized protein LOC105646185 [Jatropha curcas] |
| 20 |
Hb_004174_030 |
0.1438804484 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase IMK2 [Populus euphratica] |