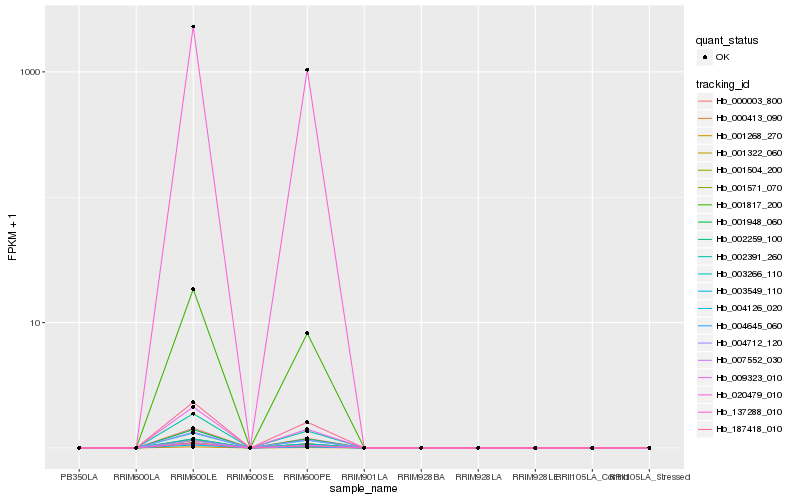
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001817_200 |
0.0 |
- |
- |
hypothetical protein CISIN_1g037825mg [Citrus sinensis] |
| 2 |
Hb_002391_260 |
0.0018795489 |
- |
- |
hypothetical protein RCOM_0017280 [Ricinus communis] |
| 3 |
Hb_001571_070 |
0.0027158729 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 4 |
Hb_000413_090 |
0.0058967474 |
- |
- |
PREDICTED: transcription factor DYT1 [Jatropha curcas] |
| 5 |
Hb_002259_100 |
0.0070099609 |
- |
- |
PREDICTED: uncharacterized protein LOC105630152 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004645_060 |
0.0119282729 |
- |
- |
PREDICTED: uncharacterized protein LOC105630948 [Jatropha curcas] |
| 7 |
Hb_001322_060 |
0.0122770231 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_000003_800 |
0.0122945854 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_137288_010 |
0.013246525 |
- |
- |
hypothetical protein B456_003G055000 [Gossypium raimondii] |
| 10 |
Hb_003266_110 |
0.0141757839 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_001948_060 |
0.0169981974 |
- |
- |
PREDICTED: glutathione S-transferase DHAR3, chloroplastic [Jatropha curcas] |
| 12 |
Hb_020479_010 |
0.018238832 |
- |
- |
cinnamyl alcohol dehydrogenase [Pyrus pyrifolia] |
| 13 |
Hb_187418_010 |
0.020687523 |
- |
- |
PREDICTED: TMV resistance protein N-like, partial [Citrus sinensis] |
| 14 |
Hb_009323_010 |
0.023339876 |
- |
- |
hypothetical protein POPTR_0008s03350g, partial [Populus trichocarpa] |
| 15 |
Hb_001504_200 |
0.0253443439 |
- |
- |
Beta-3 adrenergic receptor, putative [Theobroma cacao] |
| 16 |
Hb_007552_030 |
0.0266800523 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21064 [Jatropha curcas] |
| 17 |
Hb_004712_120 |
0.0276740214 |
- |
- |
Pectinesterase U1 precursor, putative [Ricinus communis] |
| 18 |
Hb_003549_110 |
0.0278660923 |
- |
- |
PREDICTED: uncharacterized protein LOC105647409 [Jatropha curcas] |
| 19 |
Hb_004126_020 |
0.0314947316 |
- |
- |
PREDICTED: uncharacterized protein LOC105649549 [Jatropha curcas] |
| 20 |
Hb_001268_270 |
0.0350792382 |
- |
- |
protein binding protein, putative [Ricinus communis] |