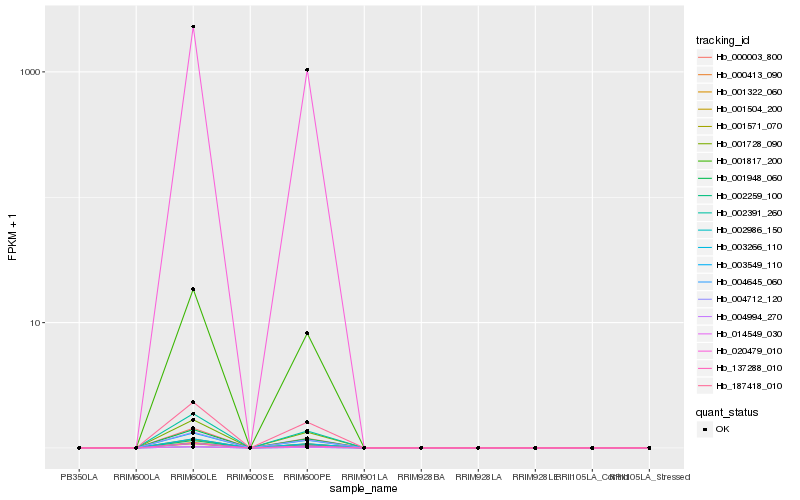
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003266_110 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_137288_010 |
0.0009293419 |
- |
- |
hypothetical protein B456_003G055000 [Gossypium raimondii] |
| 3 |
Hb_000003_800 |
0.0018813546 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001322_060 |
0.0018989181 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_001948_060 |
0.0028227356 |
- |
- |
PREDICTED: glutathione S-transferase DHAR3, chloroplastic [Jatropha curcas] |
| 6 |
Hb_020479_010 |
0.0040635448 |
- |
- |
cinnamyl alcohol dehydrogenase [Pyrus pyrifolia] |
| 7 |
Hb_187418_010 |
0.0065126396 |
- |
- |
PREDICTED: TMV resistance protein N-like, partial [Citrus sinensis] |
| 8 |
Hb_000413_090 |
0.0082793684 |
- |
- |
PREDICTED: transcription factor DYT1 [Jatropha curcas] |
| 9 |
Hb_001504_200 |
0.0111704419 |
- |
- |
Beta-3 adrenergic receptor, putative [Theobroma cacao] |
| 10 |
Hb_001571_070 |
0.0114601234 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 11 |
Hb_002391_260 |
0.0122963929 |
- |
- |
hypothetical protein RCOM_0017280 [Ricinus communis] |
| 12 |
Hb_004712_120 |
0.0135007144 |
- |
- |
Pectinesterase U1 precursor, putative [Ricinus communis] |
| 13 |
Hb_003549_110 |
0.0136928374 |
- |
- |
PREDICTED: uncharacterized protein LOC105647409 [Jatropha curcas] |
| 14 |
Hb_001817_200 |
0.0141757839 |
- |
- |
hypothetical protein CISIN_1g037825mg [Citrus sinensis] |
| 15 |
Hb_002259_100 |
0.0211847192 |
- |
- |
PREDICTED: uncharacterized protein LOC105630152 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001728_090 |
0.0216312193 |
transcription factor |
TF Family: MYB |
MYB family protein [Jatropha curcas] |
| 17 |
Hb_002986_150 |
0.0240183249 |
- |
- |
hypothetical protein glysoja_022060 [Glycine soja] |
| 18 |
Hb_004645_060 |
0.0261018938 |
- |
- |
PREDICTED: uncharacterized protein LOC105630948 [Jatropha curcas] |
| 19 |
Hb_004994_270 |
0.0333754542 |
- |
- |
PREDICTED: tRNA-specific adenosine deaminase 1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_014549_030 |
0.0342063439 |
- |
- |
receptor-kinase, putative [Ricinus communis] |