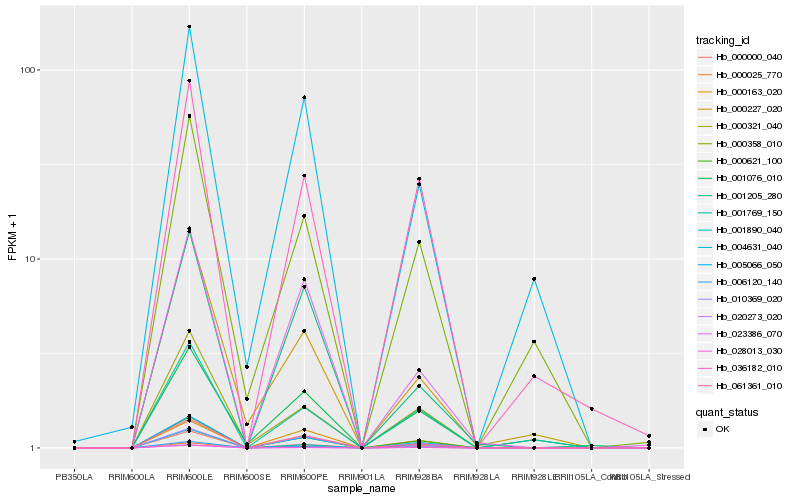
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000621_100 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_006120_140 |
0.0465388695 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 3 |
Hb_000000_040 |
0.0794809762 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075-like [Jatropha curcas] |
| 4 |
Hb_000025_770 |
0.0833301994 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 5 |
Hb_000321_040 |
0.0856163439 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 5 [Jatropha curcas] |
| 6 |
Hb_061361_010 |
0.0858631215 |
- |
- |
kinase, putative [Ricinus communis] |
| 7 |
Hb_005066_050 |
0.0887405243 |
- |
- |
- |
| 8 |
Hb_020273_020 |
0.1032710236 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 9 |
Hb_036182_010 |
0.1068987542 |
- |
- |
hypothetical protein POPTR_0473s00220g [Populus trichocarpa] |
| 10 |
Hb_010369_020 |
0.112220031 |
- |
- |
hypothetical protein JCGZ_21088 [Jatropha curcas] |
| 11 |
Hb_001890_040 |
0.1174240834 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 12 |
Hb_001769_150 |
0.1227275167 |
- |
- |
PREDICTED: uncharacterized protein LOC105633924 [Jatropha curcas] |
| 13 |
Hb_000163_020 |
0.1336107768 |
- |
- |
- |
| 14 |
Hb_028013_030 |
0.1336535668 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_023386_070 |
0.1358628769 |
- |
- |
PREDICTED: probable pectinesterase 68 [Jatropha curcas] |
| 16 |
Hb_000358_010 |
0.1444213362 |
- |
- |
PREDICTED: basic blue protein [Populus euphratica] |
| 17 |
Hb_001205_280 |
0.1447695733 |
- |
- |
PREDICTED: probable polyol transporter 6 [Jatropha curcas] |
| 18 |
Hb_000227_020 |
0.1478207702 |
- |
- |
PREDICTED: basic 7S globulin-like [Jatropha curcas] |
| 19 |
Hb_001076_010 |
0.1480261249 |
transcription factor |
TF Family: B3 |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_004631_040 |
0.1517791128 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |