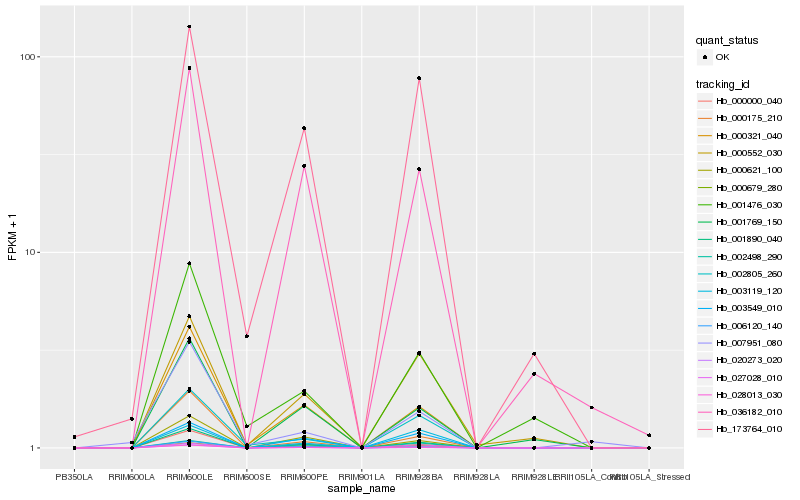
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028013_030 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_001890_040 |
0.0249514298 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 3 |
Hb_006120_140 |
0.1031907336 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 4 |
Hb_020273_020 |
0.1085486475 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 5 |
Hb_000321_040 |
0.1090122071 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 5 [Jatropha curcas] |
| 6 |
Hb_000175_210 |
0.1259271445 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 7 |
Hb_000621_100 |
0.1336535668 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003549_010 |
0.1354854676 |
- |
- |
PREDICTED: uncharacterized protein LOC105647434 [Jatropha curcas] |
| 9 |
Hb_002805_260 |
0.135727228 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_036182_010 |
0.1366576772 |
- |
- |
hypothetical protein POPTR_0473s00220g [Populus trichocarpa] |
| 11 |
Hb_000000_040 |
0.1401969297 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075-like [Jatropha curcas] |
| 12 |
Hb_027028_010 |
0.1420434722 |
- |
- |
hypothetical protein JCGZ_01141 [Jatropha curcas] |
| 13 |
Hb_001769_150 |
0.1451288493 |
- |
- |
PREDICTED: uncharacterized protein LOC105633924 [Jatropha curcas] |
| 14 |
Hb_173764_010 |
0.1577806916 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_000552_030 |
0.1617090309 |
- |
- |
glucose-6-phosphate 1-dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_003119_120 |
0.1710978052 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Nicotiana sylvestris] |
| 17 |
Hb_000679_280 |
0.1716506286 |
- |
- |
hypothetical protein POPTR_0006s27120g [Populus trichocarpa] |
| 18 |
Hb_001476_030 |
0.1812119675 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 19 |
Hb_007951_080 |
0.1835737673 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 20 |
Hb_002498_290 |
0.1874500235 |
- |
- |
PREDICTED: autophagy-related protein 8C-like isoform X2 [Elaeis guineensis] |