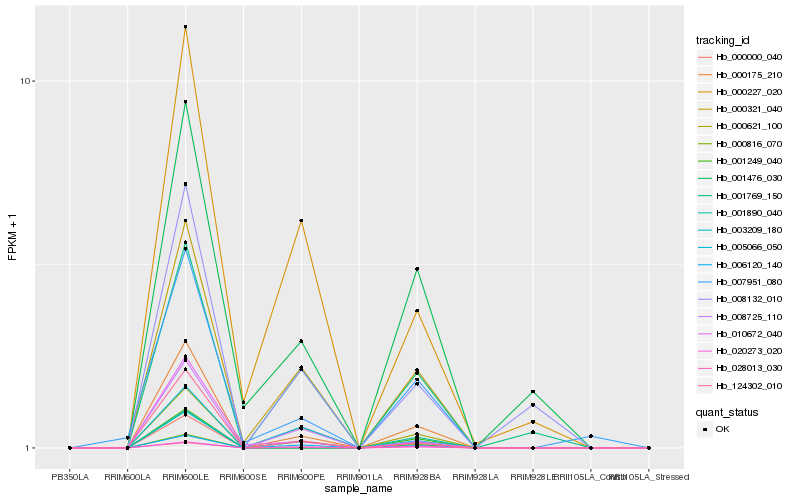
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000175_210 |
0.0 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 2 |
Hb_020273_020 |
0.0771855888 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 3 |
Hb_001890_040 |
0.1087914844 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 4 |
Hb_000000_040 |
0.1193021521 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075-like [Jatropha curcas] |
| 5 |
Hb_124302_010 |
0.1250188972 |
- |
- |
PREDICTED: serine carboxypeptidase-like [Jatropha curcas] |
| 6 |
Hb_028013_030 |
0.1259271445 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_006120_140 |
0.1314447028 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 8 |
Hb_000321_040 |
0.136026398 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 5 [Jatropha curcas] |
| 9 |
Hb_008725_110 |
0.1546146323 |
- |
- |
PREDICTED: leucine-rich repeat extensin-like protein 1 [Jatropha curcas] |
| 10 |
Hb_007951_080 |
0.1700940337 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 11 |
Hb_010672_040 |
0.1732922782 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 12 |
Hb_000621_100 |
0.1766593592 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003209_180 |
0.1828235324 |
- |
- |
Alpha-1,4 glucan phosphorylase L-2 isozyme [Morus notabilis] |
| 14 |
Hb_001769_150 |
0.1857651579 |
- |
- |
PREDICTED: uncharacterized protein LOC105633924 [Jatropha curcas] |
| 15 |
Hb_008132_010 |
0.1878866609 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Jatropha curcas] |
| 16 |
Hb_001249_040 |
0.1896518243 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 17 |
Hb_005066_050 |
0.1932666197 |
- |
- |
- |
| 18 |
Hb_000816_070 |
0.1951438166 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000227_020 |
0.1994050193 |
- |
- |
PREDICTED: basic 7S globulin-like [Jatropha curcas] |
| 20 |
Hb_001476_030 |
0.1998905593 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |