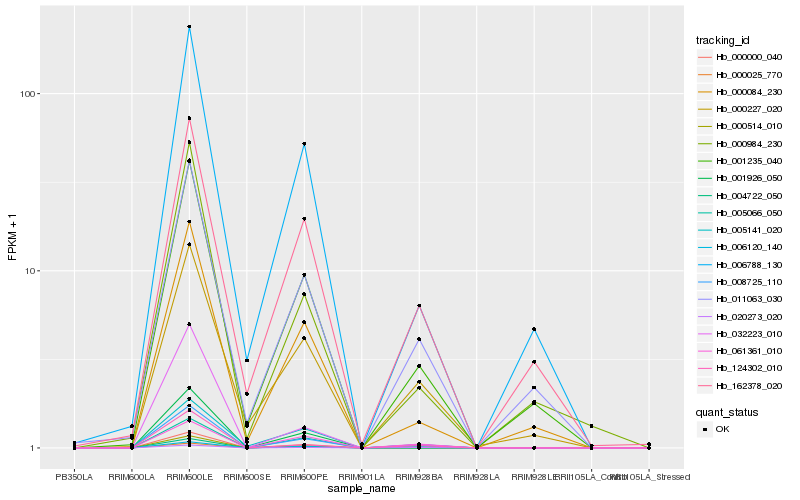
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008725_110 |
0.0 |
- |
- |
PREDICTED: leucine-rich repeat extensin-like protein 1 [Jatropha curcas] |
| 2 |
Hb_000000_040 |
0.0888716316 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075-like [Jatropha curcas] |
| 3 |
Hb_005066_050 |
0.0951696538 |
- |
- |
- |
| 4 |
Hb_001235_040 |
0.1109942454 |
- |
- |
hypothetical protein POPTR_0001s47470g [Populus trichocarpa] |
| 5 |
Hb_020273_020 |
0.1150433202 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 6 |
Hb_124302_010 |
0.1210828468 |
- |
- |
PREDICTED: serine carboxypeptidase-like [Jatropha curcas] |
| 7 |
Hb_000084_230 |
0.1248562531 |
- |
- |
hypothetical protein POPTR_0016s08030g [Populus trichocarpa] |
| 8 |
Hb_006788_130 |
0.1261316359 |
- |
- |
PREDICTED: histone H2A variant 1 [Jatropha curcas] |
| 9 |
Hb_000984_230 |
0.1340259623 |
- |
- |
- |
| 10 |
Hb_011063_030 |
0.1342001171 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_006120_140 |
0.1386676642 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 12 |
Hb_005141_020 |
0.1397576776 |
- |
- |
laccase, putative [Ricinus communis] |
| 13 |
Hb_000025_770 |
0.139988494 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 14 |
Hb_032223_010 |
0.1408629811 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 15 |
Hb_000227_020 |
0.1439036511 |
- |
- |
PREDICTED: basic 7S globulin-like [Jatropha curcas] |
| 16 |
Hb_061361_010 |
0.1454140987 |
- |
- |
kinase, putative [Ricinus communis] |
| 17 |
Hb_162378_020 |
0.1475298774 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 13-like [Populus euphratica] |
| 18 |
Hb_004722_050 |
0.152547552 |
- |
- |
hypothetical protein POPTR_0011s02010g [Populus trichocarpa] |
| 19 |
Hb_000514_010 |
0.1528635713 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA4 [Jatropha curcas] |
| 20 |
Hb_001926_050 |
0.1529156393 |
- |
- |
PREDICTED: peroxidase 19 [Jatropha curcas] |