| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
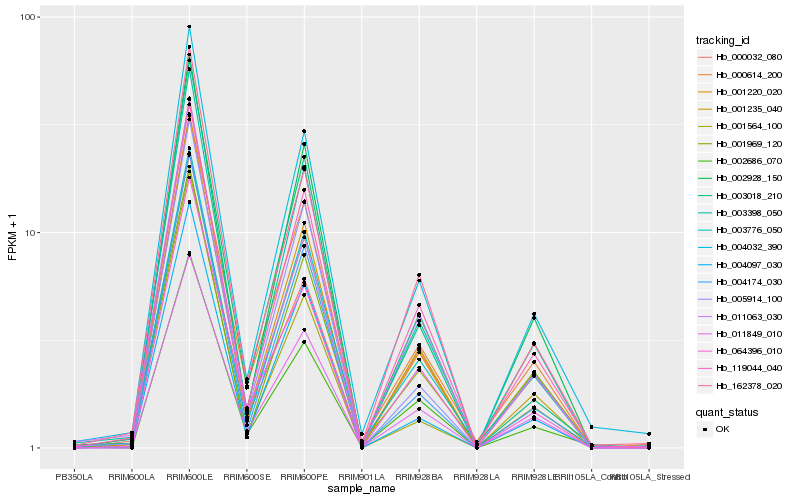
Hb_001220_020 |
0.0 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase IMK2 [Jatropha curcas] |
| 2 |
Hb_003398_050 |
0.04022693 |
- |
- |
B-type cell cycle switch protein ccs52B [Populus trichocarpa] |
| 3 |
Hb_004032_390 |
0.0452206077 |
- |
- |
CDK, putative [Ricinus communis] |
| 4 |
Hb_162378_020 |
0.0501053323 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 13-like [Populus euphratica] |
| 5 |
Hb_004174_030 |
0.0537082004 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase IMK2 [Populus euphratica] |
| 6 |
Hb_002928_150 |
0.0540716957 |
- |
- |
PREDICTED: cyclin-B2-4-like [Jatropha curcas] |
| 7 |
Hb_001235_040 |
0.0662892737 |
- |
- |
hypothetical protein POPTR_0001s47470g [Populus trichocarpa] |
| 8 |
Hb_003776_050 |
0.0679115821 |
- |
- |
PREDICTED: uncharacterized protein LOC105641219 [Jatropha curcas] |
| 9 |
Hb_004097_030 |
0.0680535493 |
- |
- |
PREDICTED: uncharacterized protein LOC105634518 [Jatropha curcas] |
| 10 |
Hb_064396_010 |
0.0704625468 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_003018_210 |
0.0712595494 |
- |
- |
PREDICTED: uncharacterized protein LOC105138721 isoform X2 [Populus euphratica] |
| 12 |
Hb_005914_100 |
0.0722280669 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000614_200 |
0.0737860343 |
- |
- |
plant mitotic spindle assembly checkpoint protein mad2, putative [Ricinus communis] |
| 14 |
Hb_119044_040 |
0.0737896628 |
- |
- |
cyclin B, putative [Ricinus communis] |
| 15 |
Hb_001969_120 |
0.0754871161 |
- |
- |
PREDICTED: plectin [Jatropha curcas] |
| 16 |
Hb_000032_080 |
0.0756356319 |
transcription factor |
TF Family: HMG |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_002686_070 |
0.076369288 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_011849_010 |
0.076993392 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 isoform X4 [Jatropha curcas] |
| 19 |
Hb_011063_030 |
0.0788257929 |
- |
- |
protein with unknown function [Ricinus communis] |
| 20 |
Hb_001564_100 |
0.0797465061 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |