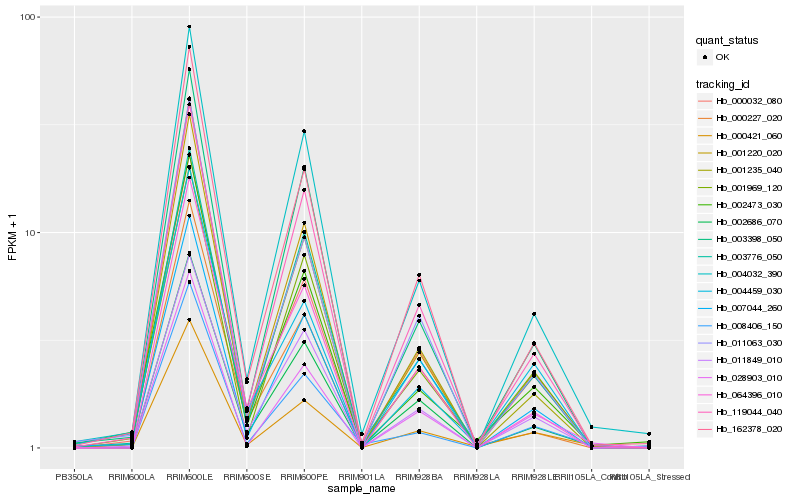
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000032_080 |
0.0 |
transcription factor |
TF Family: HMG |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_162378_020 |
0.0553185763 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 13-like [Populus euphratica] |
| 3 |
Hb_011063_030 |
0.0591012917 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_001220_020 |
0.0756356319 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase IMK2 [Jatropha curcas] |
| 5 |
Hb_064396_010 |
0.0761469801 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_002686_070 |
0.0777459881 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_007044_260 |
0.0814175001 |
- |
- |
PREDICTED: uncharacterized protein LOC105638706 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004459_030 |
0.0864847359 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000421_060 |
0.0865539218 |
- |
- |
PREDICTED: uncharacterized protein LOC105628158 [Jatropha curcas] |
| 10 |
Hb_011849_010 |
0.0887770571 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 isoform X4 [Jatropha curcas] |
| 11 |
Hb_004032_390 |
0.0888724747 |
- |
- |
CDK, putative [Ricinus communis] |
| 12 |
Hb_001235_040 |
0.0895072874 |
- |
- |
hypothetical protein POPTR_0001s47470g [Populus trichocarpa] |
| 13 |
Hb_001969_120 |
0.0911687168 |
- |
- |
PREDICTED: plectin [Jatropha curcas] |
| 14 |
Hb_119044_040 |
0.0923801219 |
- |
- |
cyclin B, putative [Ricinus communis] |
| 15 |
Hb_028903_010 |
0.0931839082 |
- |
- |
hypothetical protein JCGZ_09851 [Jatropha curcas] |
| 16 |
Hb_003398_050 |
0.09592114 |
- |
- |
B-type cell cycle switch protein ccs52B [Populus trichocarpa] |
| 17 |
Hb_002473_030 |
0.0968336999 |
- |
- |
PREDICTED: osmotic avoidance abnormal protein 3 [Jatropha curcas] |
| 18 |
Hb_003776_050 |
0.0995617788 |
- |
- |
PREDICTED: uncharacterized protein LOC105641219 [Jatropha curcas] |
| 19 |
Hb_000227_020 |
0.1000059323 |
- |
- |
PREDICTED: basic 7S globulin-like [Jatropha curcas] |
| 20 |
Hb_008406_150 |
0.1035863661 |
- |
- |
PREDICTED: mitotic checkpoint serine/threonine-protein kinase BUB1 isoform X1 [Jatropha curcas] |