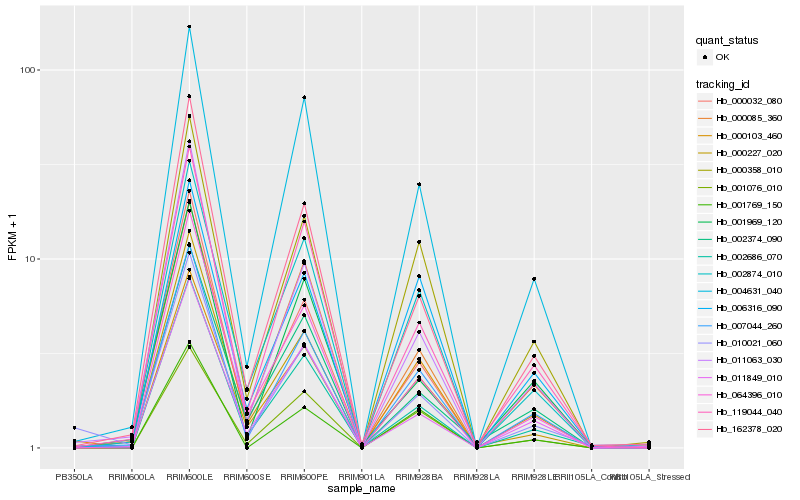
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007044_260 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638706 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000358_010 |
0.0631261449 |
- |
- |
PREDICTED: basic blue protein [Populus euphratica] |
| 3 |
Hb_004631_040 |
0.0749310622 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 4 |
Hb_002874_010 |
0.0808161593 |
- |
- |
PREDICTED: replication protein A 32 kDa subunit A isoform X1 [Jatropha curcas] |
| 5 |
Hb_000032_080 |
0.0814175001 |
transcription factor |
TF Family: HMG |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_162378_020 |
0.0840090666 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 13-like [Populus euphratica] |
| 7 |
Hb_002686_070 |
0.085364795 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_119044_040 |
0.0868964999 |
- |
- |
cyclin B, putative [Ricinus communis] |
| 9 |
Hb_011063_030 |
0.0894793746 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_001076_010 |
0.0975379649 |
transcription factor |
TF Family: B3 |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_000085_360 |
0.0983983372 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 12 |
Hb_064396_010 |
0.1017894213 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_011849_010 |
0.1041936998 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 isoform X4 [Jatropha curcas] |
| 14 |
Hb_001769_150 |
0.1060932108 |
- |
- |
PREDICTED: uncharacterized protein LOC105633924 [Jatropha curcas] |
| 15 |
Hb_000227_020 |
0.1063101165 |
- |
- |
PREDICTED: basic 7S globulin-like [Jatropha curcas] |
| 16 |
Hb_006316_090 |
0.1075627842 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 17 |
Hb_002374_090 |
0.1079418561 |
- |
- |
PREDICTED: cyclin-A1-1 [Jatropha curcas] |
| 18 |
Hb_001969_120 |
0.1084694724 |
- |
- |
PREDICTED: plectin [Jatropha curcas] |
| 19 |
Hb_000103_460 |
0.1089844577 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 20 |
Hb_010021_060 |
0.1092857467 |
- |
- |
PREDICTED: kinesin-related protein 11 isoform X1 [Jatropha curcas] |