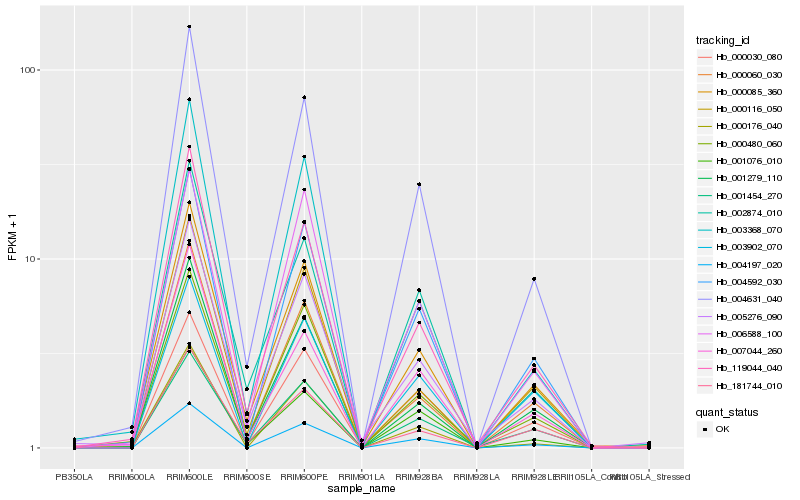
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004197_020 |
0.0 |
- |
- |
Anthocyanin 5-aromatic acyltransferase, putative [Ricinus communis] |
| 2 |
Hb_004592_030 |
0.0326577915 |
- |
- |
unknown [Lotus japonicus] |
| 3 |
Hb_004631_040 |
0.0716652486 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 4 |
Hb_000176_040 |
0.0846799814 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHR3 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000030_080 |
0.0863749055 |
- |
- |
PREDICTED: uncharacterized protein LOC105632640 [Jatropha curcas] |
| 6 |
Hb_003902_070 |
0.093353198 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000480_060 |
0.096894451 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 8 |
Hb_000085_360 |
0.1004335911 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 9 |
Hb_001076_010 |
0.1023064946 |
transcription factor |
TF Family: B3 |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_001279_110 |
0.103033614 |
- |
- |
PREDICTED: uncharacterized protein LOC105633338 isoform X4 [Jatropha curcas] |
| 11 |
Hb_119044_040 |
0.1068058585 |
- |
- |
cyclin B, putative [Ricinus communis] |
| 12 |
Hb_006588_100 |
0.1077441472 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000060_030 |
0.1089687495 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1-like [Jatropha curcas] |
| 14 |
Hb_005276_090 |
0.1133988147 |
- |
- |
PREDICTED: uncharacterized protein LOC105649997 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001454_270 |
0.1152628463 |
- |
- |
PREDICTED: condensin-2 complex subunit D3 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000116_050 |
0.1161298343 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin-2-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_003368_070 |
0.1171631068 |
- |
- |
PREDICTED: uncharacterized protein LOC105109387 [Populus euphratica] |
| 18 |
Hb_007044_260 |
0.120935596 |
- |
- |
PREDICTED: uncharacterized protein LOC105638706 isoform X1 [Jatropha curcas] |
| 19 |
Hb_181744_010 |
0.1233837116 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 20 |
Hb_002874_010 |
0.1245604713 |
- |
- |
PREDICTED: replication protein A 32 kDa subunit A isoform X1 [Jatropha curcas] |