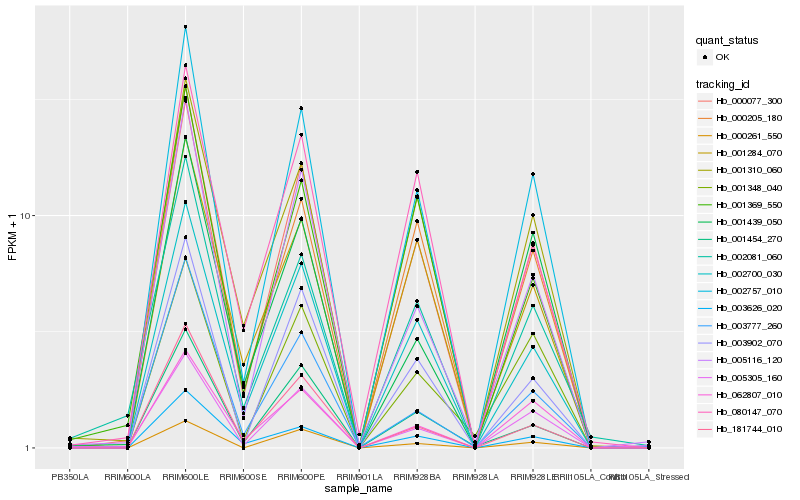
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002757_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0013s10580g [Populus trichocarpa] |
| 2 |
Hb_000077_300 |
0.0484465409 |
- |
- |
PREDICTED: expansin-B3-like [Jatropha curcas] |
| 3 |
Hb_002700_030 |
0.0808401541 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 4 |
Hb_003902_070 |
0.0823641927 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000205_180 |
0.0866008272 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 6 |
Hb_062807_010 |
0.1004500529 |
- |
- |
hypothetical protein JCGZ_15896 [Jatropha curcas] |
| 7 |
Hb_005116_120 |
0.1007279187 |
- |
- |
PREDICTED: uncharacterized protein LOC105649997 isoform X2 [Jatropha curcas] |
| 8 |
Hb_003626_020 |
0.103716368 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At5g24080 [Jatropha curcas] |
| 9 |
Hb_001310_060 |
0.1047325445 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 10 |
Hb_005305_160 |
0.1085801026 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_181744_010 |
0.1123810172 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 12 |
Hb_000261_550 |
0.1129343378 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2-like [Jatropha curcas] |
| 13 |
Hb_001454_270 |
0.1152348037 |
- |
- |
PREDICTED: condensin-2 complex subunit D3 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001348_040 |
0.1156076434 |
- |
- |
hypothetical protein VITISV_006651 [Vitis vinifera] |
| 15 |
Hb_002081_060 |
0.117349368 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 16 |
Hb_001284_070 |
0.1175406139 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003777_260 |
0.1197396722 |
- |
- |
PREDICTED: uncharacterized protein LOC105640927 isoform X4 [Jatropha curcas] |
| 18 |
Hb_001439_050 |
0.1242869061 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |
| 19 |
Hb_080147_070 |
0.1254562048 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 20 |
Hb_001369_550 |
0.1254877546 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |