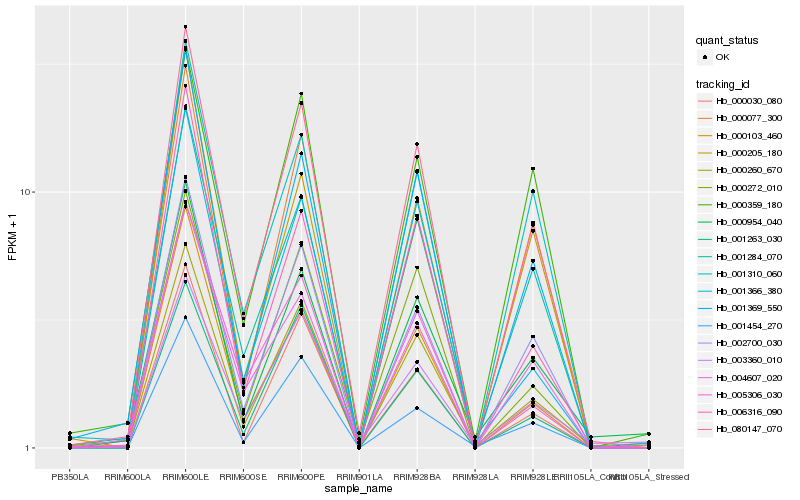
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_080147_070 |
0.0 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 2 |
Hb_001284_070 |
0.0650785725 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002700_030 |
0.0674880162 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 4 |
Hb_000260_670 |
0.0708131165 |
- |
- |
PREDICTED: serine/threonine-protein kinase TIO isoform X1 [Jatropha curcas] |
| 5 |
Hb_001369_550 |
0.0751395657 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 6 |
Hb_001310_060 |
0.0795965503 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 7 |
Hb_004607_020 |
0.0841515339 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 8 |
Hb_000272_010 |
0.0948765653 |
- |
- |
PREDICTED: DNA replication licensing factor MCM5 [Jatropha curcas] |
| 9 |
Hb_000077_300 |
0.0973367087 |
- |
- |
PREDICTED: expansin-B3-like [Jatropha curcas] |
| 10 |
Hb_000205_180 |
0.1027347786 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 11 |
Hb_003360_010 |
0.1032804907 |
- |
- |
Bipolar kinesin KRP-130, putative [Ricinus communis] |
| 12 |
Hb_000030_080 |
0.1070417253 |
- |
- |
PREDICTED: uncharacterized protein LOC105632640 [Jatropha curcas] |
| 13 |
Hb_000359_180 |
0.1082126359 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000954_040 |
0.1089851655 |
- |
- |
PREDICTED: metal tolerance protein 4 [Jatropha curcas] |
| 15 |
Hb_005306_030 |
0.109070764 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 [Jatropha curcas] |
| 16 |
Hb_001366_380 |
0.1138901176 |
- |
- |
PREDICTED: pectin acetylesterase 6-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_006316_090 |
0.1167920375 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 18 |
Hb_001263_030 |
0.1171959548 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 19 |
Hb_001454_270 |
0.1193989763 |
- |
- |
PREDICTED: condensin-2 complex subunit D3 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000103_460 |
0.1211651151 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |