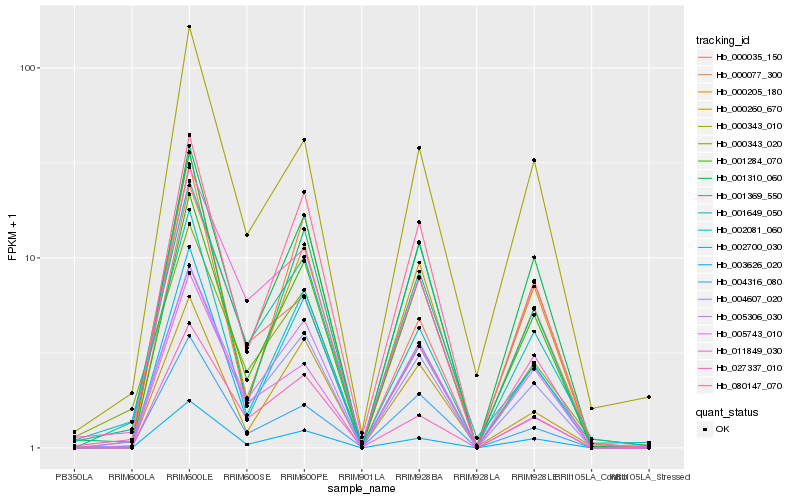
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004607_020 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 2 |
Hb_001284_070 |
0.0782373976 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001310_060 |
0.0806442505 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 4 |
Hb_080147_070 |
0.0841515339 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 5 |
Hb_003626_020 |
0.0873387289 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At5g24080 [Jatropha curcas] |
| 6 |
Hb_002700_030 |
0.0909643306 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 7 |
Hb_001369_550 |
0.0997825125 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 8 |
Hb_001649_050 |
0.1012619919 |
- |
- |
Shaggy-like kinase 13 isoform 1 [Theobroma cacao] |
| 9 |
Hb_000205_180 |
0.1018167202 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 10 |
Hb_004316_080 |
0.1022367589 |
- |
- |
PREDICTED: uncharacterized protein LOC104602212 [Nelumbo nucifera] |
| 11 |
Hb_005306_030 |
0.1068332369 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 [Jatropha curcas] |
| 12 |
Hb_000343_010 |
0.1090099345 |
- |
- |
F3F9.11 [Arabidopsis thaliana] |
| 13 |
Hb_027337_010 |
0.1108480342 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 14 |
Hb_002081_060 |
0.1121503614 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 15 |
Hb_000260_670 |
0.1178602066 |
- |
- |
PREDICTED: serine/threonine-protein kinase TIO isoform X1 [Jatropha curcas] |
| 16 |
Hb_011849_030 |
0.1179606792 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005743_010 |
0.1181656865 |
- |
- |
RING/U-box superfamily protein, putative [Theobroma cacao] |
| 18 |
Hb_000343_020 |
0.1196952542 |
- |
- |
PREDICTED: putative Peroxidase 48 [Jatropha curcas] |
| 19 |
Hb_000077_300 |
0.1204075322 |
- |
- |
PREDICTED: expansin-B3-like [Jatropha curcas] |
| 20 |
Hb_000035_150 |
0.1235005438 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |