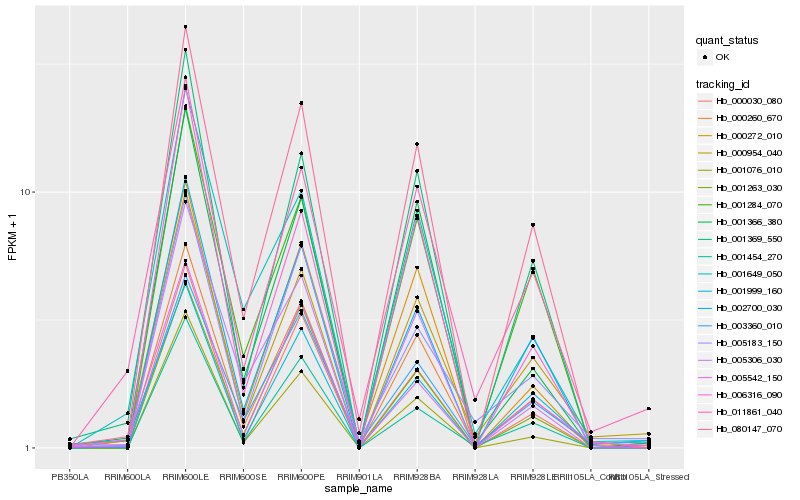
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000260_670 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase TIO isoform X1 [Jatropha curcas] |
| 2 |
Hb_080147_070 |
0.0708131165 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 3 |
Hb_000954_040 |
0.0756608366 |
- |
- |
PREDICTED: metal tolerance protein 4 [Jatropha curcas] |
| 4 |
Hb_000272_010 |
0.0771930766 |
- |
- |
PREDICTED: DNA replication licensing factor MCM5 [Jatropha curcas] |
| 5 |
Hb_003360_010 |
0.083983672 |
- |
- |
Bipolar kinesin KRP-130, putative [Ricinus communis] |
| 6 |
Hb_001366_380 |
0.0873389932 |
- |
- |
PREDICTED: pectin acetylesterase 6-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_002700_030 |
0.0902820818 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 8 |
Hb_001369_550 |
0.0931999548 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 9 |
Hb_000030_080 |
0.1037081558 |
- |
- |
PREDICTED: uncharacterized protein LOC105632640 [Jatropha curcas] |
| 10 |
Hb_005183_150 |
0.1070475849 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 11 |
Hb_005306_030 |
0.1093347057 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 [Jatropha curcas] |
| 12 |
Hb_001263_030 |
0.1097139571 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 13 |
Hb_001284_070 |
0.1100380636 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001649_050 |
0.1101944101 |
- |
- |
Shaggy-like kinase 13 isoform 1 [Theobroma cacao] |
| 15 |
Hb_001999_160 |
0.1135883499 |
- |
- |
hypothetical protein JCGZ_08488 [Jatropha curcas] |
| 16 |
Hb_001076_010 |
0.1140724736 |
transcription factor |
TF Family: B3 |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_001454_270 |
0.1141885986 |
- |
- |
PREDICTED: condensin-2 complex subunit D3 isoform X2 [Jatropha curcas] |
| 18 |
Hb_011861_040 |
0.1144994274 |
- |
- |
PREDICTED: pheromone-processing carboxypeptidase KEX1-like [Jatropha curcas] |
| 19 |
Hb_005542_150 |
0.1149222429 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 20 |
Hb_006316_090 |
0.1171806653 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |