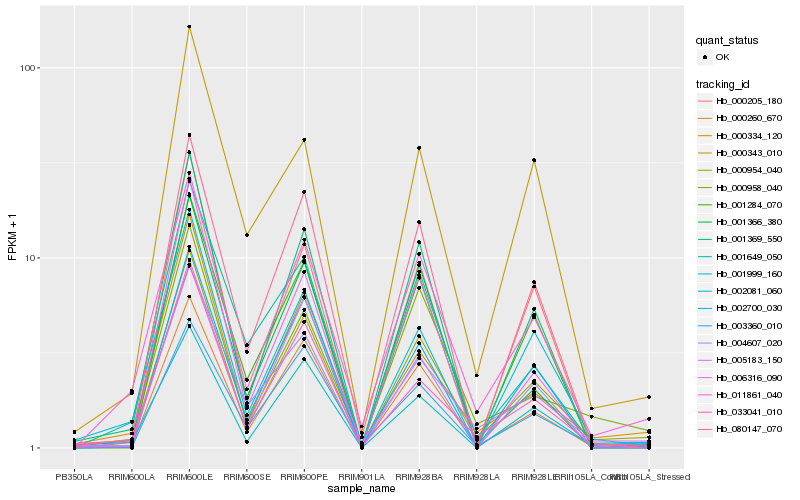
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000954_040 |
0.0 |
- |
- |
PREDICTED: metal tolerance protein 4 [Jatropha curcas] |
| 2 |
Hb_000260_670 |
0.0756608366 |
- |
- |
PREDICTED: serine/threonine-protein kinase TIO isoform X1 [Jatropha curcas] |
| 3 |
Hb_011861_040 |
0.0925255994 |
- |
- |
PREDICTED: pheromone-processing carboxypeptidase KEX1-like [Jatropha curcas] |
| 4 |
Hb_001369_550 |
0.1026447398 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 5 |
Hb_080147_070 |
0.1089851655 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 6 |
Hb_002700_030 |
0.1094284036 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 7 |
Hb_001999_160 |
0.1126560508 |
- |
- |
hypothetical protein JCGZ_08488 [Jatropha curcas] |
| 8 |
Hb_033041_010 |
0.1128668981 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2-like [Populus euphratica] |
| 9 |
Hb_005183_150 |
0.1131454596 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 10 |
Hb_000205_180 |
0.1160170028 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 11 |
Hb_003360_010 |
0.1179264509 |
- |
- |
Bipolar kinesin KRP-130, putative [Ricinus communis] |
| 12 |
Hb_001284_070 |
0.1211847532 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002081_060 |
0.1221832401 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 14 |
Hb_000334_120 |
0.1227117663 |
- |
- |
PREDICTED: DNA topoisomerase 2 [Jatropha curcas] |
| 15 |
Hb_006316_090 |
0.125371741 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 16 |
Hb_004607_020 |
0.1254636723 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 17 |
Hb_000958_040 |
0.1274100624 |
- |
- |
PREDICTED: DNA topoisomerase 2 [Jatropha curcas] |
| 18 |
Hb_001649_050 |
0.1277058576 |
- |
- |
Shaggy-like kinase 13 isoform 1 [Theobroma cacao] |
| 19 |
Hb_001366_380 |
0.1279181389 |
- |
- |
PREDICTED: pectin acetylesterase 6-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_000343_010 |
0.1324734268 |
- |
- |
F3F9.11 [Arabidopsis thaliana] |