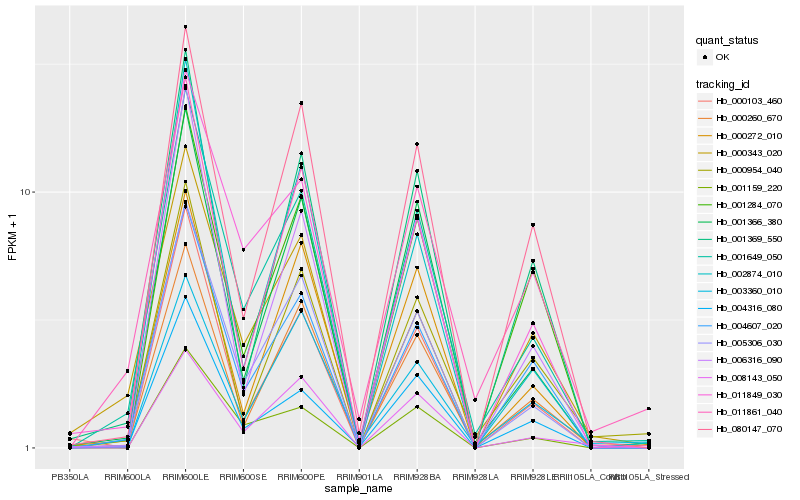
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001649_050 |
0.0 |
- |
- |
Shaggy-like kinase 13 isoform 1 [Theobroma cacao] |
| 2 |
Hb_005306_030 |
0.0849002691 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 [Jatropha curcas] |
| 3 |
Hb_011849_030 |
0.0965008464 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004607_020 |
0.1012619919 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 5 |
Hb_001366_380 |
0.1038330513 |
- |
- |
PREDICTED: pectin acetylesterase 6-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_004316_080 |
0.1083375988 |
- |
- |
PREDICTED: uncharacterized protein LOC104602212 [Nelumbo nucifera] |
| 7 |
Hb_000260_670 |
0.1101944101 |
- |
- |
PREDICTED: serine/threonine-protein kinase TIO isoform X1 [Jatropha curcas] |
| 8 |
Hb_080147_070 |
0.1217673791 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 9 |
Hb_001159_220 |
0.1243148796 |
- |
- |
ESC, putative [Ricinus communis] |
| 10 |
Hb_006316_090 |
0.1273091825 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 11 |
Hb_000954_040 |
0.1277058576 |
- |
- |
PREDICTED: metal tolerance protein 4 [Jatropha curcas] |
| 12 |
Hb_011861_040 |
0.129840771 |
- |
- |
PREDICTED: pheromone-processing carboxypeptidase KEX1-like [Jatropha curcas] |
| 13 |
Hb_000103_460 |
0.1313911884 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 14 |
Hb_000272_010 |
0.1339418801 |
- |
- |
PREDICTED: DNA replication licensing factor MCM5 [Jatropha curcas] |
| 15 |
Hb_008143_050 |
0.1361081288 |
- |
- |
PREDICTED: cytochrome P450 710A1-like [Jatropha curcas] |
| 16 |
Hb_000343_020 |
0.1363170974 |
- |
- |
PREDICTED: putative Peroxidase 48 [Jatropha curcas] |
| 17 |
Hb_002874_010 |
0.1363303182 |
- |
- |
PREDICTED: replication protein A 32 kDa subunit A isoform X1 [Jatropha curcas] |
| 18 |
Hb_001369_550 |
0.1367186395 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 19 |
Hb_001284_070 |
0.1374090253 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003360_010 |
0.1387825025 |
- |
- |
Bipolar kinesin KRP-130, putative [Ricinus communis] |